You are browsing environment: FUNGIDB
CAZyme Information: ATY64919.1
You are here: Home > Sequence: ATY64919.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cordyceps militaris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Cordycipitaceae; Cordyceps; Cordyceps militaris | |||||||||||
| CAZyme ID | ATY64919.1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | Glycoside family 35 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5115132; End:5118608 Strand: + | |||||||||||
Full Sequence Download help
| MKFLWGALVA LSALSATLAA ETTHAPGSFS YNRTDFLLNG QPFQIIGGQM DPQRIPPEYW | 60 |
| THRLKMARAM GLNTIFSYLY WNLHESRPGA WDFSGRNDVA RFFRLAQQEG LQVVLRPGPY | 120 |
| ICGERDWGGF PAWLSQVPGM AVRQNNRPFL DAAKSYLDRL GKELGQLQIT QGGPILMTQL | 180 |
| ENEYGSFGTD KTYLAALAAM LRDNFDVFLY TNDGGGQSYL EGGQLHGVLA VIDGDSQSGF | 240 |
| AARDKYVTDP TSLGPQLNGE YYISWIDQWG SDYPHQQIAG SQADVAKAVA DLDWTLAGGY | 300 |
| SFSIYMFHGG TNFGFENGGI RDDGPLAAMT TSYDYGAPLD ESGRPTDVYF RLRDMIQKYV | 360 |
| PKGSIPSVPA MPARAAVPEF QLRPAAALFD LQGRPTRQAS DPVSMDALGQ AYGYVLYQHT | 420 |
| VATDVAGNVA IGDGARDRAI IYVNGVRSGV VDTIYKTPST VSVTLRKGDK LQILVENLGR | 480 |
| VDVRQRLREQ VKGIVGHVSV GGTVLTNWCM HSIPLDTLPA GLDGKKTHVV RQKDGPVFYT | 540 |
| GSFDMPAGAA ADPSGDTFLA VPKGIKGVLW VNGVNMGRYW TVGPQQSLTH NTVDTSSTLT | 600 |
| LAMSRPQTPP HEPRYNVHVA PTTISQLIRT AFPNIELVSS SELTSHRGYN NRLYLLTVRR | 660 |
| RGGPSCVFRD TDAAERELVL KANGRFFLAD KVQNEVGCLQ VLGQYCPAIP TPTVFAWSEE | 720 |
| GHDVCLASPA GPEIKNVTLA IPDGEKRHGG WILMSRLPGA PLSVCDLDEV SRLDIMRQLA | 780 |
| GVTASWRTNI PAQRYIGNIQ FHQSVHASEP DFAIVKNSGP RPQDLVVRGM LVDELRITTP | 840 |
| ITSVTEQYTR KLEQKLTLLE TSDTYRPNRH LAPEIRRFVA ETLPRLTKQQ PSHFVFTHYD | 900 |
| LSPRNILVGG SPPQISGIVD FEFAGFFPPV EEFLNDAVGN EGDWPDHLYA AYLAELEARG | 960 |
| VATPAAGIGA AEWETARCLE RVADNVAPWW LPGKYTGSAL EEQFAKSAAE LRENMRKLS | 1019 |
Enzyme Prediction help
| EC | 3.2.1.23:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 36 | 357 | 1.1e-97 | 0.993485342019544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 6.50e-113 | 35 | 356 | 1 | 315 | Glycosyl hydrolases family 35. |
| 166698 | PLN03059 | 2.75e-55 | 8 | 580 | 9 | 659 | beta-galactosidase; Provisional |
| 224786 | GanA | 2.28e-32 | 29 | 594 | 1 | 579 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 396281 | APH | 8.40e-08 | 667 | 960 | 14 | 232 | Phosphotransferase enzyme family. This family consists of bacterial antibiotic resistance proteins, which confer resistance to various aminoglycosides they include: aminoglycoside 3'-phosphotransferase or kanamycin kinase / neomycin-kanamycin phosphotransferase and streptomycin 3''-kinase or streptomycin 3''-phosphotransferase. The aminoglycoside phosphotransferases inactivate aminoglycoside antibiotics via phosphorylation. This family also includes homoserine kinase. This family is related to fructosamine kinase pfam03881. |
| 270690 | APH_ChoK_like | 1.28e-07 | 875 | 929 | 94 | 145 | Aminoglycoside 3'-phosphotransferase and Choline Kinase family. This family is composed of APH, ChoK, ethanolamine kinase (ETNK), macrolide 2'-phosphotransferase (MPH2'), an unusual homoserine kinase, and uncharacterized proteins with similarity to the N-terminal domain of acyl-CoA dehydrogenase 10 (ACAD10). The members of this family catalyze the transfer of the gamma-phosphoryl group from ATP (or CTP) to small molecule substrates such as aminoglycosides, macrolides, choline, ethanolamine, and homoserine. Phosphorylation of the antibiotics, aminoglycosides and macrolides, leads to their inactivation and to bacterial antibiotic resistance. Phosphorylation of choline, ethanolamine, and homoserine serves as precursors to the synthesis of important biological compounds, such as the major phospholipids, phosphatidylcholine and phosphatidylethanolamine and the amino acids, threonine, methionine, and isoleucine. The APH/ChoK family is part of a larger superfamily that includes the catalytic domains of other kinases, such as the typical serine/threonine/tyrosine protein kinases (PKs), RIO kinases, actin-fragmin kinase (AFK), and phosphoinositide 3-kinase (PI3K). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATY64919.1|GH35 | 0.0 | 1 | 1019 | 1 | 1019 |
| UNI19525.1|GH35 | 5.14e-274 | 8 | 588 | 10 | 588 |
| QLI69147.1|GH35 | 3.81e-265 | 3 | 588 | 11 | 595 |
| QKX60376.1|GH35 | 2.04e-253 | 8 | 588 | 14 | 602 |
| QRD02051.1|GH35 | 1.83e-180 | 25 | 588 | 33 | 589 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KDV_A | 1.02e-104 | 21 | 588 | 10 | 586 | Chain A, Beta-galactosidase [Mus musculus],7KDV_C Chain C, Beta-galactosidase [Mus musculus],7KDV_E Chain E, Beta-galactosidase [Mus musculus],7KDV_G Chain G, Beta-galactosidase [Mus musculus],7KDV_I Chain I, Beta-galactosidase [Mus musculus],7KDV_K Chain K, Beta-galactosidase [Mus musculus] |
| 3THC_A | 1.97e-103 | 31 | 588 | 13 | 578 | Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
| 3WEZ_A | 2.64e-103 | 17 | 588 | 23 | 602 | Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_B Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_C Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_D Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WF0_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF1_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF2_A Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_B Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_C Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_D Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens] |
| 3WF3_A | 1.38e-102 | 17 | 588 | 23 | 602 | Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_B Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_C Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_D Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF4_A Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_B Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_C Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_D Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens] |
| 6EON_A | 8.38e-101 | 27 | 588 | 26 | 574 | Galactanase BT0290 [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P48982|BGAL_XANMN | 4.97e-110 | 8 | 588 | 9 | 571 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
| sp|Q9TRY9|BGAL_CANLF | 1.98e-104 | 1 | 588 | 7 | 602 | Beta-galactosidase OS=Canis lupus familiaris OX=9615 GN=GLB1 PE=1 SV=3 |
| sp|O19015|BGAL_FELCA | 2.82e-104 | 31 | 588 | 37 | 603 | Beta-galactosidase OS=Felis catus OX=9685 GN=GLB1 PE=2 SV=1 |
| sp|P23780|BGAL_MOUSE | 2.24e-103 | 21 | 588 | 27 | 603 | Beta-galactosidase OS=Mus musculus OX=10090 GN=Glb1 PE=1 SV=1 |
| sp|Q60HF6|BGAL_MACFA | 1.51e-102 | 30 | 588 | 35 | 601 | Beta-galactosidase OS=Macaca fascicularis OX=9541 GN=GLB1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
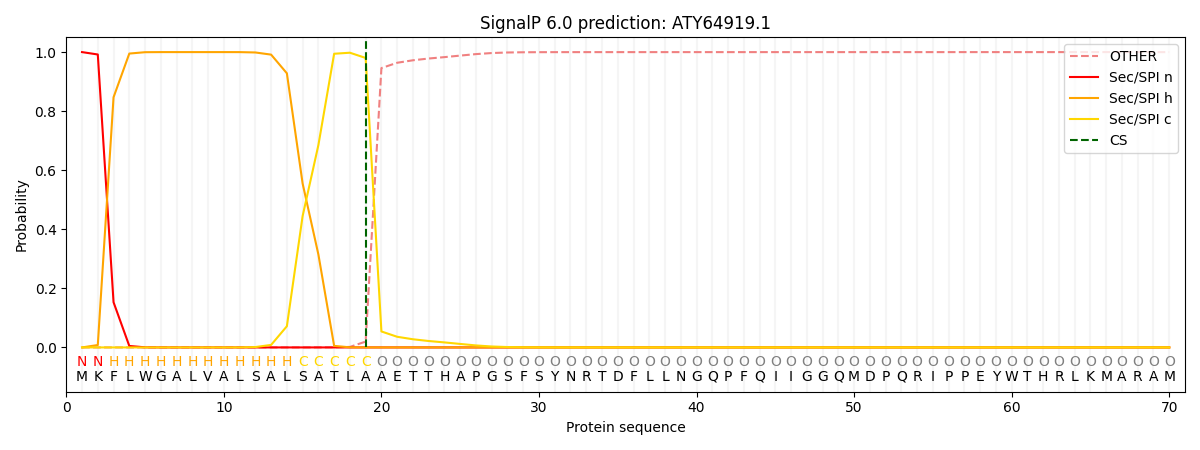
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000296 | 0.999674 | CS pos: 19-20. Pr: 0.9802 |
