You are browsing environment: FUNGIDB
CAZyme Information: ATY59959.1
You are here: Home > Sequence: ATY59959.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
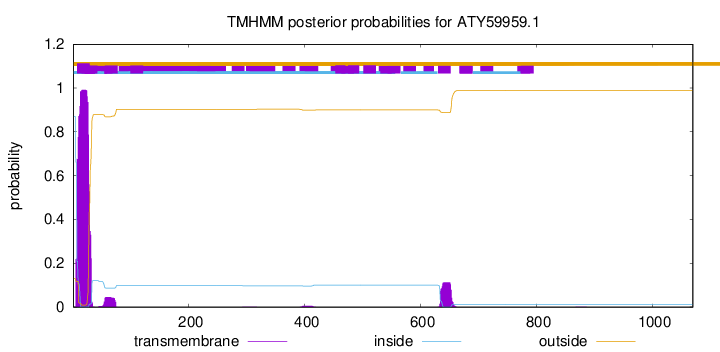
TMHMM annotations
Basic Information help
| Species | Cordyceps militaris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Cordycipitaceae; Cordyceps; Cordyceps militaris | |||||||||||
| CAZyme ID | ATY59959.1 | |||||||||||
| CAZy Family | GT90 | |||||||||||
| CAZyme Description | ubiquitin C-terminal | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 239127 | Peptidase_C19F | 1.58e-43 | 46 | 625 | 1 | 240 | A subfamily of Peptidase C19. Peptidase C19 contains ubiquitinyl hydrolases. They are intracellular peptidases that remove ubiquitin molecules from polyubiquinated peptides by cleavage of isopeptide bonds. They hydrolyze bonds involving the carboxyl group of the C-terminal Gly residue of ubiquitin. The purpose of the de-ubiquitination is thought to be editing of the ubiquitin conjugates, which could rescue them from degradation, as well as recycling of the ubiquitin. The ubiquitin/proteasome system is responsible for most protein turnover in the mammalian cell, and with over 50 members, family C19 is one of the largest families of peptidases in the human genome. |
| 395355 | UCH | 1.63e-33 | 45 | 466 | 1 | 275 | Ubiquitin carboxyl-terminal hydrolase. |
| 239072 | Peptidase_C19 | 1.67e-21 | 156 | 466 | 16 | 218 | Peptidase C19 contains ubiquitinyl hydrolases. They are intracellular peptidases that remove ubiquitin molecules from polyubiquinated peptides by cleavage of isopeptide bonds. They hydrolyse bonds involving the carboxyl group of the C-terminal Gly residue of ubiquitin The purpose of the de-ubiquitination is thought to be editing of the ubiquitin conjugates, which could rescue them from degradation, as well as recycling of the ubiquitin. The ubiquitin/proteasome system is responsible for most protein turnover in the mammalian cell, and with over 50 members, family C19 is one of the largest families of peptidases in the human genome. |
| 239139 | Peptidase_C19R | 4.84e-17 | 272 | 468 | 46 | 200 | A subfamily of peptidase C19. Peptidase C19 contains ubiquitinyl hydrolases. They are intracellular peptidases that remove ubiquitin molecules from polyubiquinated peptides by cleavage of isopeptide bonds. They hydrolyze bonds involving the carboxyl group of the C-terminal Gly residue of ubiquitin. The purpose of the de-ubiquitination is thought to be editing of the ubiquitin conjugates, which could rescue them from degradation, as well as recycling of the ubiquitin. The ubiquitin/proteasome system is responsible for most protein turnover in the mammalian cell, and with over 50 members, family C19 is one of the largest families of peptidases in the human genome. |
| 227820 | COG5533 | 4.74e-16 | 44 | 466 | 71 | 381 | Ubiquitin C-terminal hydrolase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1070 | 1 | 1070 | |
| 1.73e-152 | 626 | 1070 | 4 | 454 | |
| 7.66e-152 | 624 | 1069 | 13 | 494 | |
| 1.89e-151 | 626 | 1069 | 4 | 453 | |
| 1.89e-151 | 626 | 1069 | 4 | 453 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.91e-11 | 45 | 475 | 4 | 327 | Crystal structure of covalent ubiquitin-USP21 complex [Homo sapiens],3I3T_C Crystal structure of covalent ubiquitin-USP21 complex [Homo sapiens],3I3T_E Crystal structure of covalent ubiquitin-USP21 complex [Homo sapiens],3I3T_G Crystal structure of covalent ubiquitin-USP21 complex [Homo sapiens] |
|
| 4.33e-11 | 45 | 475 | 17 | 340 | Structure of USP21 in complex with linear diubiquitin-aldehyde [Homo sapiens],2Y5B_E Structure of USP21 in complex with linear diubiquitin-aldehyde [Homo sapiens] |
|
| 4.41e-11 | 45 | 475 | 23 | 346 | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor [Homo sapiens],3MTN_C Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.31e-145 | 1 | 628 | 1 | 621 | Ubiquitin carboxyl-terminal hydrolase 16 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=ubp16 PE=3 SV=1 |
|
| 1.88e-17 | 20 | 465 | 25 | 414 | Ubiquitin carboxyl-terminal hydrolase 16 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=UBP16 PE=1 SV=1 |
|
| 4.27e-10 | 45 | 475 | 212 | 535 | Ubiquitin carboxyl-terminal hydrolase 21 OS=Homo sapiens OX=9606 GN=USP21 PE=1 SV=1 |
|
| 2.94e-09 | 45 | 475 | 212 | 535 | Ubiquitin carboxyl-terminal hydrolase 21 OS=Rattus norvegicus OX=10116 GN=Usp21 PE=2 SV=1 |
|
| 2.94e-09 | 45 | 475 | 212 | 535 | Ubiquitin carboxyl-terminal hydrolase 21 OS=Bos taurus OX=9913 GN=USP21 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.801781 | 0.198233 |