You are browsing environment: FUNGIDB
CAZyme Information: ATY58494.1
You are here: Home > Sequence: ATY58494.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
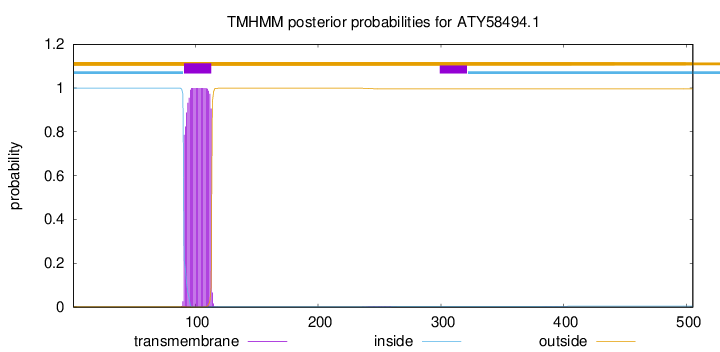
TMHMM annotations
Basic Information help
| Species | Cordyceps militaris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Cordycipitaceae; Cordyceps; Cordyceps militaris | |||||||||||
| CAZyme ID | ATY58494.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | cellulase (glycosyl hydrolase family 5) | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 160 | 472 | 1e-147 | 0.9873015873015873 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 2.15e-24 | 161 | 469 | 1 | 270 | Cellulase (glycosyl hydrolase family 5). |
| 225344 | BglC | 1.39e-16 | 170 | 469 | 47 | 361 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 223021 | PHA03247 | 7.76e-04 | 9 | 93 | 273 | 359 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 505 | 1 | 505 | |
| 3.28e-204 | 79 | 505 | 84 | 505 | |
| 5.37e-203 | 94 | 505 | 95 | 505 | |
| 3.30e-202 | 75 | 505 | 72 | 497 | |
| 2.43e-199 | 75 | 505 | 70 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.42e-39 | 155 | 489 | 8 | 338 | Chain A, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus],1VRX_B Chain B, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus] |
|
| 2.49e-38 | 155 | 489 | 8 | 338 | Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus],1ECE_B Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus] |
|
| 2.76e-21 | 164 | 489 | 55 | 396 | Functional analysis of hyperthermophilic endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],3AXX_B Functional analysis of hyperthermophilic endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],3AXX_C Functional analysis of hyperthermophilic endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3] |
|
| 3.26e-21 | 164 | 489 | 22 | 363 | The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_B The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_C The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3W6L_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
|
| 6.66e-21 | 164 | 489 | 55 | 396 | Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_A Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_B Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_C Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.18e-36 | 155 | 489 | 49 | 379 | Endoglucanase E1 OS=Acidothermus cellulolyticus (strain ATCC 43068 / DSM 8971 / 11B) OX=351607 GN=Acel_0614 PE=1 SV=1 |
|
| 7.75e-33 | 155 | 505 | 41 | 391 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
|
| 6.00e-26 | 151 | 504 | 29 | 399 | Glycosyl hydrolase 5 family protein OS=Chamaecyparis obtusa OX=13415 PE=1 SV=1 |
|
| 2.55e-24 | 153 | 489 | 46 | 400 | Endoglucanase D OS=Cellulomonas fimi OX=1708 GN=cenD PE=3 SV=1 |
|
| 4.14e-20 | 153 | 435 | 630 | 936 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000046 | 0.000000 |