You are browsing environment: FUNGIDB
CAZyme Information: ATEG_08623-t26_1-p1
You are here: Home > Sequence: ATEG_08623-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus terreus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus terreus | |||||||||||
| CAZyme ID | ATEG_08623-t26_1-p1 | |||||||||||
| CAZy Family | GT109 | |||||||||||
| CAZyme Description | predicted protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 98 | 429 | 1.4e-88 | 0.888268156424581 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 1.99e-37 | 167 | 409 | 63 | 328 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 227170 | COG4833 | 2.97e-15 | 145 | 399 | 48 | 309 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.18e-157 | 61 | 435 | 8 | 378 | |
| 1.30e-156 | 64 | 435 | 11 | 378 | |
| 1.30e-156 | 64 | 435 | 11 | 378 | |
| 5.17e-116 | 58 | 433 | 6 | 384 | |
| 5.17e-116 | 58 | 433 | 6 | 384 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.65e-22 | 145 | 405 | 48 | 307 | Chain A, Lin0763 protein [Listeria innocua] |
|
| 3.80e-17 | 183 | 399 | 86 | 306 | Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 3.93e-17 | 183 | 399 | 89 | 309 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
|
| 4.69e-17 | 183 | 399 | 107 | 327 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 1.31e-16 | 183 | 399 | 120 | 340 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.33e-12 | 187 | 373 | 114 | 315 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=DCW1 PE=3 SV=1 |
|
| 3.67e-08 | 187 | 373 | 122 | 325 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
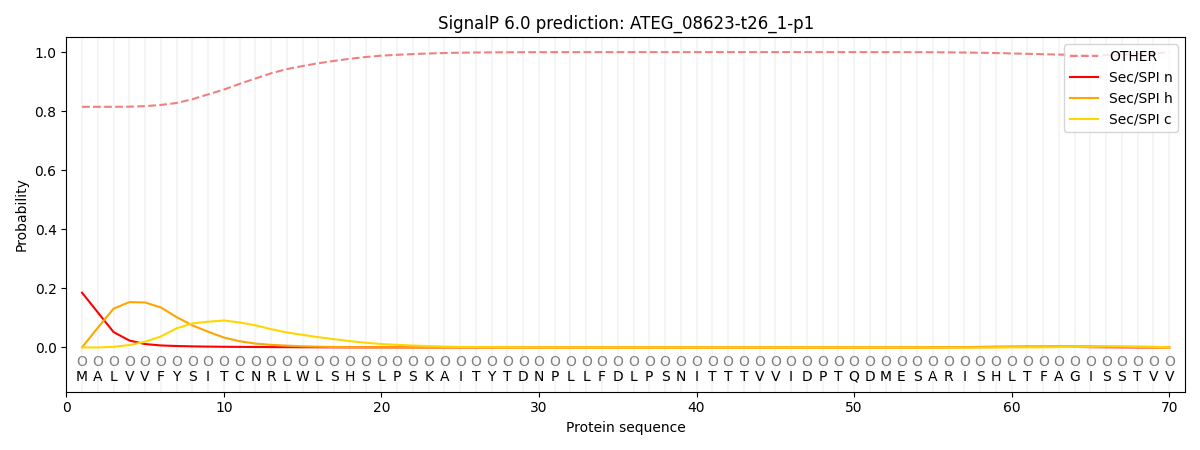
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.828159 | 0.171847 |
