You are browsing environment: FUNGIDB
CAZyme Information: ATEG_03427-t26_1-p1
You are here: Home > Sequence: ATEG_03427-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus terreus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus terreus | |||||||||||
| CAZyme ID | ATEG_03427-t26_1-p1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.22:58 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 121 | 274 | 2.3e-47 | 0.5676855895196506 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 269893 | GH27 | 5.47e-111 | 28 | 290 | 1 | 271 | glycosyl hydrolase family 27 (GH27). GH27 enzymes occur in eukaryotes, prokaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-N-acetylgalactosaminidase, and 3-alpha-isomalto-dextranase. All GH27 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH27 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| 166449 | PLN02808 | 8.51e-107 | 20 | 391 | 24 | 386 | alpha-galactosidase |
| 178295 | PLN02692 | 1.92e-99 | 20 | 386 | 48 | 406 | alpha-galactosidase |
| 177874 | PLN02229 | 3.63e-96 | 20 | 394 | 55 | 423 | alpha-galactosidase |
| 374582 | Melibiase_2 | 2.06e-66 | 27 | 290 | 1 | 284 | Alpha galactosidase A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.97e-143 | 13 | 438 | 14 | 494 | |
| 3.97e-143 | 13 | 438 | 14 | 494 | |
| 2.25e-142 | 13 | 438 | 14 | 494 | |
| 4.52e-142 | 13 | 438 | 14 | 494 | |
| 9.04e-142 | 13 | 438 | 14 | 494 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.40e-119 | 22 | 437 | 24 | 471 | Chain A, Alpha-galactosidase 1 [Saccharomyces cerevisiae],3LRL_A Chain A, Alpha-galactosidase 1 [Saccharomyces cerevisiae] |
|
| 1.18e-117 | 22 | 437 | 24 | 471 | Chain A, Alpha-galactosidase 1 [Saccharomyces cerevisiae],3LRM_B Chain B, Alpha-galactosidase 1 [Saccharomyces cerevisiae],3LRM_C Chain C, Alpha-galactosidase 1 [Saccharomyces cerevisiae],3LRM_D Chain D, Alpha-galactosidase 1 [Saccharomyces cerevisiae] |
|
| 2.62e-103 | 21 | 316 | 2 | 322 | Crystal structure of alpha-galactosidase I from Mortierella vinacea [Umbelopsis vinacea] |
|
| 9.36e-81 | 22 | 314 | 3 | 296 | Chain A, alpha-galactosidase [Oryza sativa] |
|
| 2.54e-77 | 20 | 313 | 1 | 295 | Nicotiana benthamiana alpha-galactosidase [Nicotiana benthamiana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.50e-125 | 22 | 434 | 23 | 467 | Alpha-galactosidase OS=Lachancea cidri OX=29831 GN=MEL PE=3 SV=1 |
|
| 3.66e-122 | 22 | 434 | 24 | 468 | Alpha-galactosidase OS=Saccharomyces pastorianus (strain ATCC 76529 / Carlsberg bottom yeast no.1 / CBS 1513 / CLIB 176 / NBRC 1167 / NCYC 396 / NRRL Y-12693) OX=1073566 GN=MEL PE=3 SV=1 |
|
| 1.65e-120 | 22 | 434 | 24 | 468 | Alpha-galactosidase OS=Saccharomyces mikatae OX=114525 GN=MEL PE=3 SV=1 |
|
| 4.66e-120 | 22 | 434 | 24 | 468 | Alpha-galactosidase 6 OS=Saccharomyces cerevisiae OX=4932 GN=MEL6 PE=3 SV=1 |
|
| 2.63e-119 | 22 | 434 | 24 | 468 | Alpha-galactosidase 5 OS=Saccharomyces cerevisiae OX=4932 GN=MEL5 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
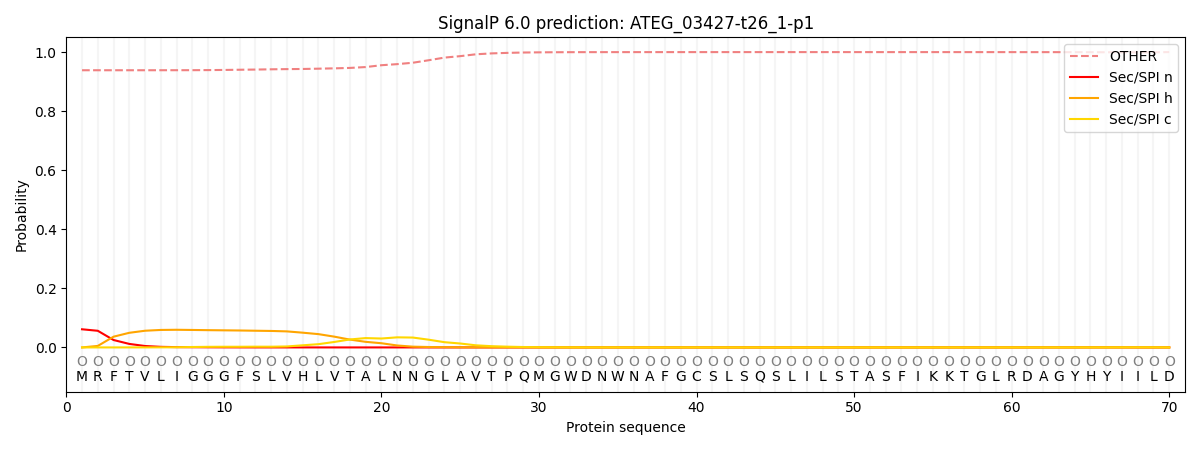
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.943229 | 0.056779 |
