You are browsing environment: FUNGIDB
CAZyme Information: ATEG_00972-t26_1-p1
You are here: Home > Sequence: ATEG_00972-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus terreus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus terreus | |||||||||||
| CAZyme ID | ATEG_00972-t26_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | predicted protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH79 | 49 | 362 | 3.8e-62 | 0.7076923076923077 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.39e-83 | 40 | 360 | 45 | 372 | |
| 9.07e-83 | 40 | 360 | 45 | 372 | |
| 9.07e-83 | 40 | 360 | 45 | 372 | |
| 4.66e-82 | 40 | 359 | 42 | 368 | |
| 1.00e-81 | 40 | 360 | 45 | 372 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.80e-26 | 50 | 349 | 26 | 309 | Chain A, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase [Fusarium oxysporum],7DFS_A Chain A, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase [Fusarium oxysporum] |
|
| 3.17e-12 | 28 | 366 | 7 | 347 | beta-glucuronidase with an activity-based probe (N-acyl cyclophellitol aziridine) bound [Acidobacterium capsulatum],5G0Q_A beta-glucuronidase with an activity-based probe (N-alkyl cyclophellitol aziridine) bound [Acidobacterium capsulatum] |
|
| 3.25e-12 | 28 | 366 | 7 | 347 | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum [Acidobacterium capsulatum ATCC 51196],3VNZ_A Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with D-glucuronic acid [Acidobacterium capsulatum ATCC 51196],3VO0_A Crystal structure of beta-glucuronidase from Acidobacterium capsulatum covalent-bonded with 2-deoxy-2-fluoro-D-glucuronic acid [Acidobacterium capsulatum ATCC 51196] |
|
| 7.82e-12 | 28 | 366 | 27 | 367 | A glycoside hydrolase mutant with an unreacted activity based probe bound [Acidobacterium capsulatum] |
|
| 9.23e-09 | 40 | 370 | 23 | 319 | Crystallographic structure of a bacterial heparanase [Burkholderia pseudomallei],5BWI_B Crystallographic structure of a bacterial heparanase [Burkholderia pseudomallei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.39e-37 | 33 | 364 | 39 | 382 | Beta-glucuronidase OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=An02g11890 PE=1 SV=1 |
|
| 1.24e-33 | 26 | 369 | 16 | 379 | Beta-glucuronidase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=NCU00937 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
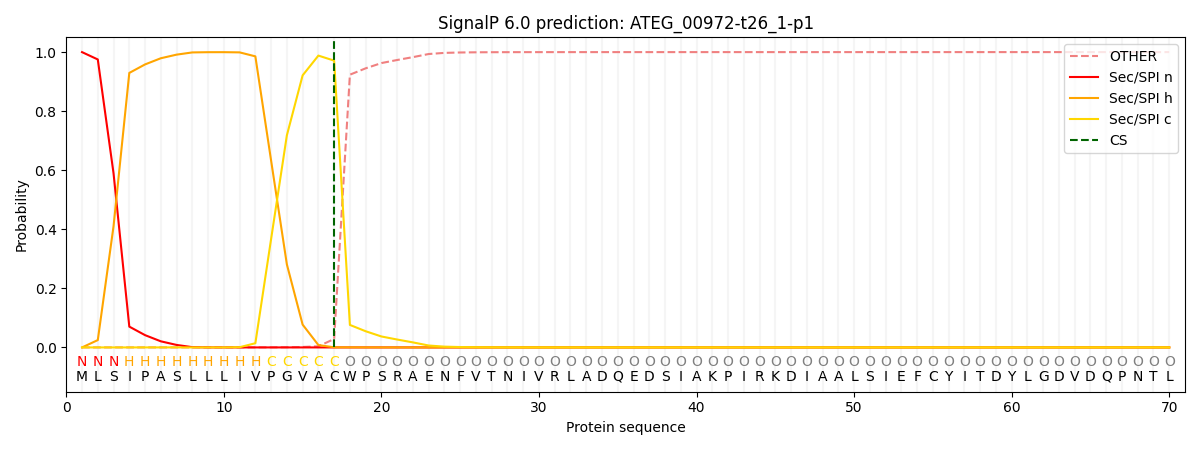
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000647 | 0.999340 | CS pos: 17-18. Pr: 0.9711 |
