You are browsing environment: FUNGIDB
CAZyme Information: ATCC64974_99900-t41_1-p1
Basic Information
help
| Species |
Aspergillus niger
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger
|
| CAZyme ID |
ATCC64974_99900-t41_1-p1
|
| CAZy Family |
GT66 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AnigerN402ATCC64974 |
11187 |
N/A |
0 |
11187
|
|
| Gene Location |
No EC number prediction in ATCC64974_99900-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
25 |
719 |
3.1e-66 |
0.7851941747572816 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 407266
|
Glyco_hydro_106 |
1.66e-12 |
251 |
687 |
439 |
868 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
4 |
951 |
2 |
998 |
| 0.0 |
4 |
951 |
2 |
998 |
| 0.0 |
31 |
744 |
53 |
800 |
| 0.0 |
12 |
948 |
11 |
999 |
| 0.0 |
25 |
947 |
18 |
1018 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 3.18e-19 |
251 |
942 |
503 |
1139 |
Structure of Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
ATCC64974_99900-t41_1-p1 has no Swissprot hit.
This protein is predicted as SP
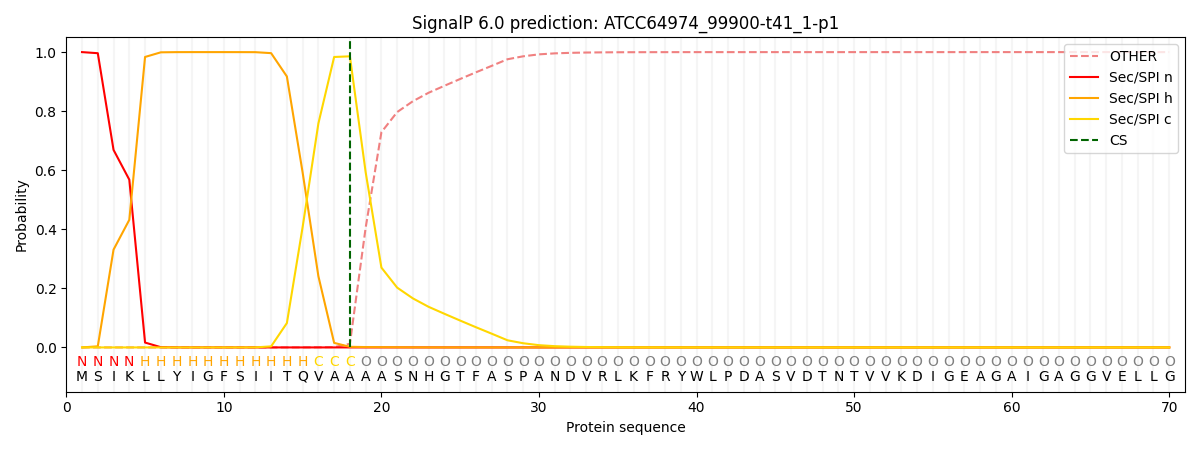
| Other |
SP_Sec_SPI |
CS Position |
| 0.000333 |
0.999629 |
CS pos: 18-19. Pr: 0.9857 |
There is no transmembrane helices in ATCC64974_99900-t41_1-p1.

