You are browsing environment: FUNGIDB
CAZyme Information: ASPZODRAFT_129029-t33_1-p1
You are here: Home > Sequence: ASPZODRAFT_129029-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Penicilliopsis zonata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Penicilliopsis; Penicilliopsis zonata | |||||||||||
| CAZyme ID | ASPZODRAFT_129029-t33_1-p1 | |||||||||||
| CAZy Family | CBM48 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.23:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 32 | 355 | 1.4e-86 | 0.993485342019544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 2.50e-96 | 31 | 354 | 1 | 315 | Glycosyl hydrolases family 35. |
| 166698 | PLN03059 | 2.83e-39 | 27 | 345 | 32 | 329 | beta-galactosidase; Provisional |
| 224786 | GanA | 1.62e-35 | 25 | 628 | 1 | 595 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 1.74e-05 | 48 | 205 | 4 | 165 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.38e-236 | 6 | 649 | 7 | 636 | |
| 3.09e-225 | 13 | 649 | 23 | 650 | |
| 2.39e-220 | 10 | 647 | 15 | 641 | |
| 8.74e-214 | 3 | 603 | 4 | 588 | |
| 6.78e-191 | 25 | 649 | 26 | 640 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.97e-112 | 31 | 628 | 23 | 583 | Chain A, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
|
| 3.85e-105 | 20 | 624 | 4 | 599 | Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
|
| 7.15e-105 | 20 | 624 | 28 | 623 | Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_B Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_C Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_D Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WF0_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF1_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF2_A Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_B Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_C Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_D Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens] |
|
| 1.98e-104 | 20 | 624 | 28 | 623 | Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_B Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_C Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF3_D Crystal structure of human beta-galactosidase mutant I51T in complex with Galactose [Homo sapiens],3WF4_A Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_B Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_C Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens],3WF4_D Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ [Homo sapiens] |
|
| 1.71e-98 | 16 | 624 | 9 | 607 | Chain A, Beta-galactosidase [Mus musculus],7KDV_C Chain C, Beta-galactosidase [Mus musculus],7KDV_E Chain E, Beta-galactosidase [Mus musculus],7KDV_G Chain G, Beta-galactosidase [Mus musculus],7KDV_I Chain I, Beta-galactosidase [Mus musculus],7KDV_K Chain K, Beta-galactosidase [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.24e-105 | 16 | 624 | 23 | 622 | Beta-galactosidase OS=Homo sapiens OX=9606 GN=GLB1 PE=1 SV=2 |
|
| 1.73e-104 | 6 | 605 | 12 | 573 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
|
| 2.90e-104 | 16 | 624 | 23 | 622 | Beta-galactosidase OS=Macaca fascicularis OX=9541 GN=GLB1 PE=2 SV=1 |
|
| 3.84e-103 | 16 | 624 | 23 | 622 | Beta-galactosidase OS=Pongo abelii OX=9601 GN=GLB1 PE=2 SV=1 |
|
| 6.16e-102 | 16 | 624 | 22 | 621 | Beta-galactosidase OS=Bos taurus OX=9913 GN=GLB1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
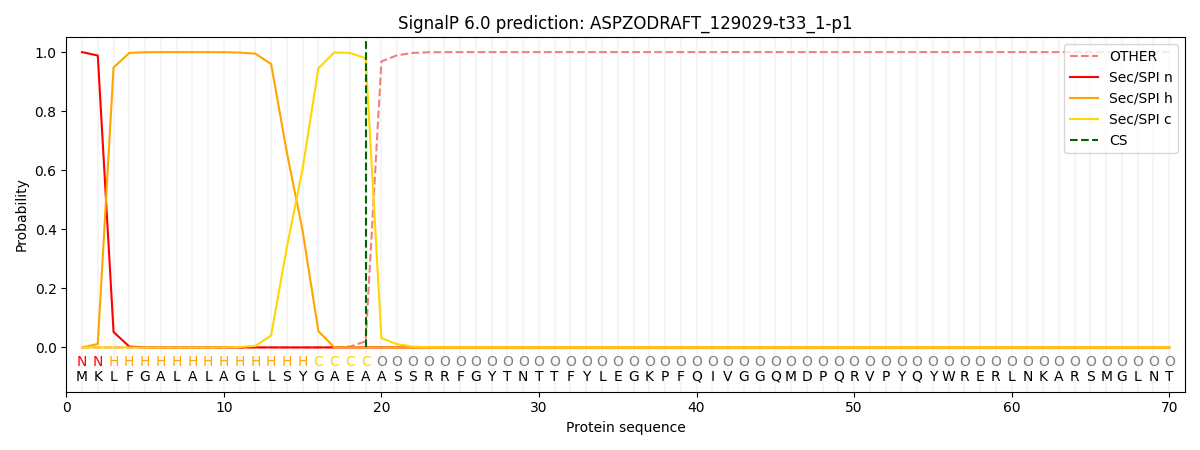
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000271 | 0.999710 | CS pos: 19-20. Pr: 0.9794 |
