You are browsing environment: FUNGIDB
CAZyme Information: ASPWEDRAFT_77488-t33_1-p1
You are here: Home > Sequence: ASPWEDRAFT_77488-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus wentii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus wentii | |||||||||||
| CAZyme ID | ASPWEDRAFT_77488-t33_1-p1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.74:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 27 | 202 | 1.2e-42 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 7.40e-59 | 27 | 203 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.95e-100 | 1 | 203 | 12 | 213 | |
| 1.25e-95 | 1 | 203 | 10 | 212 | |
| 1.02e-94 | 1 | 203 | 10 | 212 | |
| 1.02e-94 | 1 | 203 | 10 | 212 | |
| 1.20e-94 | 1 | 203 | 11 | 216 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.49e-91 | 8 | 203 | 1 | 196 | Crystal structure of Aspergillus oryzae cutinase [Aspergillus oryzae] |
|
| 4.35e-85 | 20 | 203 | 4 | 187 | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon [Aspergillus oryzae] |
|
| 2.16e-83 | 8 | 203 | 1 | 198 | Chain A, cutinase [Malbranchea cinnamomea] |
|
| 3.61e-63 | 10 | 203 | 2 | 200 | Glomerella cingulata apo cutinase [Colletotrichum gloeosporioides],3DD5_A Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_B Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_C Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_D Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_E Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_F Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_G Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DD5_H Glomerella cingulata E600-cutinase complex [Colletotrichum gloeosporioides],3DEA_A Glomerella cingulata PETFP-cutinase complex [Colletotrichum gloeosporioides],3DEA_B Glomerella cingulata PETFP-cutinase complex [Colletotrichum gloeosporioides] |
|
| 7.66e-56 | 25 | 203 | 14 | 192 | Humicola insolens cutinase [Humicola insolens],4OYY_B Humicola insolens cutinase [Humicola insolens],4OYY_C Humicola insolens cutinase [Humicola insolens],4OYY_D Humicola insolens cutinase [Humicola insolens],4OYY_E Humicola insolens cutinase [Humicola insolens],4OYY_F Humicola insolens cutinase [Humicola insolens],4OYY_G Humicola insolens cutinase [Humicola insolens],4OYY_H Humicola insolens cutinase [Humicola insolens],4OYY_I Humicola insolens cutinase [Humicola insolens],4OYY_J Humicola insolens cutinase [Humicola insolens],4OYY_K Humicola insolens cutinase [Humicola insolens],4OYY_L Humicola insolens cutinase [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.68e-103 | 1 | 203 | 8 | 211 | Probable cutinase 1 OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=ACLA_081180 PE=3 SV=1 |
|
| 3.68e-102 | 1 | 203 | 10 | 209 | Probable cutinase 1 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=ATEG_08433 PE=3 SV=1 |
|
| 3.46e-101 | 1 | 203 | 12 | 213 | Cutinase 1 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cut1 PE=2 SV=1 |
|
| 9.25e-101 | 1 | 203 | 8 | 211 | Probable cutinase 1 OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=NFIA_084890 PE=3 SV=1 |
|
| 1.31e-100 | 1 | 203 | 8 | 211 | Probable cutinase 1 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_025250 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
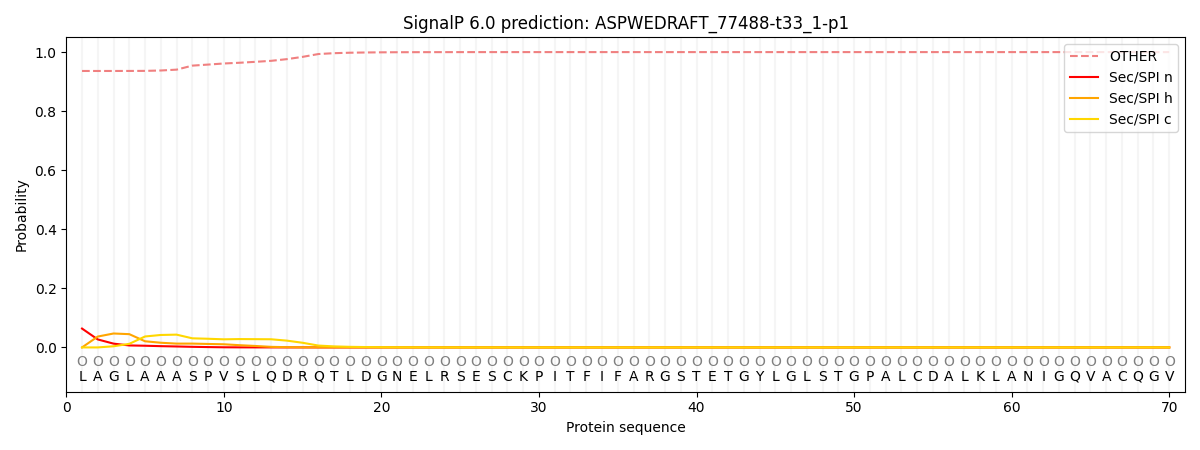
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.937509 | 0.062500 |
