You are browsing environment: FUNGIDB
CAZyme Information: ASPWEDRAFT_53926-t33_1-p1
You are here: Home > Sequence: ASPWEDRAFT_53926-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus wentii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus wentii | |||||||||||
| CAZyme ID | ASPWEDRAFT_53926-t33_1-p1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224995 | MmsB | 4.41e-53 | 461 | 756 | 3 | 286 | 3-hydroxyisobutyrate dehydrogenase or related beta-hydroxyacid dehydrogenase [Lipid transport and metabolism]. |
| 397486 | NAD_binding_2 | 1.48e-31 | 460 | 617 | 1 | 149 | NAD binding domain of 6-phosphogluconate dehydrogenase. The NAD binding domain of 6-phosphogluconate dehydrogenase adopts a Rossmann fold. |
| 340817 | GT1_Gtf-like | 2.32e-24 | 6 | 444 | 3 | 397 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 185358 | PRK15461 | 1.00e-19 | 461 | 762 | 4 | 293 | sulfolactaldehyde 3-reductase. |
| 185019 | PRK15059 | 4.41e-16 | 461 | 755 | 3 | 283 | 2-hydroxy-3-oxopropionate reductase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.76e-264 | 1 | 765 | 1 | 829 | |
| 4.49e-146 | 1 | 435 | 1 | 460 | |
| 8.17e-146 | 1 | 435 | 1 | 460 | |
| 8.17e-146 | 1 | 435 | 1 | 460 | |
| 6.53e-140 | 1 | 437 | 1 | 464 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.25e-16 | 459 | 761 | 2 | 286 | Structure of TT368 protein from Thermus Thermophilus HB8 [Thermus thermophilus HB8],1WP4_B Structure of TT368 protein from Thermus Thermophilus HB8 [Thermus thermophilus HB8],1WP4_C Structure of TT368 protein from Thermus Thermophilus HB8 [Thermus thermophilus HB8],1WP4_D Structure of TT368 protein from Thermus Thermophilus HB8 [Thermus thermophilus HB8] |
|
| 3.53e-15 | 459 | 761 | 2 | 286 | Structure of hydroxyisobutyrate dehydrogenase from thermus thermophilus HB8 [Thermus thermophilus HB8],2CVZ_B Structure of hydroxyisobutyrate dehydrogenase from thermus thermophilus HB8 [Thermus thermophilus HB8],2CVZ_C Structure of hydroxyisobutyrate dehydrogenase from thermus thermophilus HB8 [Thermus thermophilus HB8],2CVZ_D Structure of hydroxyisobutyrate dehydrogenase from thermus thermophilus HB8 [Thermus thermophilus HB8] |
|
| 2.61e-14 | 461 | 761 | 4 | 292 | Crystal structure of SLA Reductase YihU from E. Coli [Escherichia coli],6SM7_B Crystal structure of SLA Reductase YihU from E. Coli [Escherichia coli],6SM7_C Crystal structure of SLA Reductase YihU from E. Coli [Escherichia coli],6SM7_D Crystal structure of SLA Reductase YihU from E. Coli [Escherichia coli],6SMY_A Crystal structure of SLA Reductase YihU from E. Coli with NADH and product DHPS [Escherichia coli K-12],6SMY_B Crystal structure of SLA Reductase YihU from E. Coli with NADH and product DHPS [Escherichia coli K-12],6SMY_C Crystal structure of SLA Reductase YihU from E. Coli with NADH and product DHPS [Escherichia coli K-12],6SMY_D Crystal structure of SLA Reductase YihU from E. Coli with NADH and product DHPS [Escherichia coli K-12],6SMZ_A Crystal structure of SLA Reductase YihU from E. Coli in complex with NADH [Escherichia coli K-12],6SMZ_B Crystal structure of SLA Reductase YihU from E. Coli in complex with NADH [Escherichia coli K-12],6SMZ_C Crystal structure of SLA Reductase YihU from E. Coli in complex with NADH [Escherichia coli K-12],6SMZ_D Crystal structure of SLA Reductase YihU from E. Coli in complex with NADH [Escherichia coli K-12] |
|
| 2.64e-13 | 459 | 758 | 8 | 296 | The crystal structure of Bacillus cereus 3-hydroxyisobutyrate dehydrogenase in complex with NAD [Bacillus cereus ATCC 14579],5JE8_B The crystal structure of Bacillus cereus 3-hydroxyisobutyrate dehydrogenase in complex with NAD [Bacillus cereus ATCC 14579],5JE8_C The crystal structure of Bacillus cereus 3-hydroxyisobutyrate dehydrogenase in complex with NAD [Bacillus cereus ATCC 14579],5JE8_D The crystal structure of Bacillus cereus 3-hydroxyisobutyrate dehydrogenase in complex with NAD [Bacillus cereus ATCC 14579] |
|
| 2.34e-12 | 458 | 752 | 1 | 282 | Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens],3PEF_B Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens],3PEF_C Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens],3PEF_D Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens],3PEF_E Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens],3PEF_F Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens],3PEF_G Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens],3PEF_H Crystal structure of gamma-hydroxybutyrate dehydrogenase from Geobacter metallireducens in complex with NADP+ [Geobacter metallireducens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.84e-86 | 6 | 418 | 8 | 436 | Glycosyltransferase buaB OS=Aspergillus burnettii OX=2508778 GN=buaB PE=3 SV=1 |
|
| 7.25e-67 | 5 | 420 | 7 | 466 | Glycosyltransferase sdnJ OS=Sordaria araneosa OX=573841 GN=sdnJ PE=1 SV=1 |
|
| 7.16e-19 | 461 | 757 | 4 | 288 | Uncharacterized oxidoreductase Sfri_1503 OS=Shewanella frigidimarina (strain NCIMB 400) OX=318167 GN=Sfri_1503 PE=3 SV=2 |
|
| 3.26e-17 | 461 | 756 | 3 | 286 | Uncharacterized oxidoreductase YfjR OS=Bacillus subtilis (strain 168) OX=224308 GN=yfjR PE=3 SV=2 |
|
| 1.22e-13 | 461 | 761 | 4 | 292 | 3-sulfolactaldehyde reductase OS=Shigella flexneri OX=623 GN=yihU PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
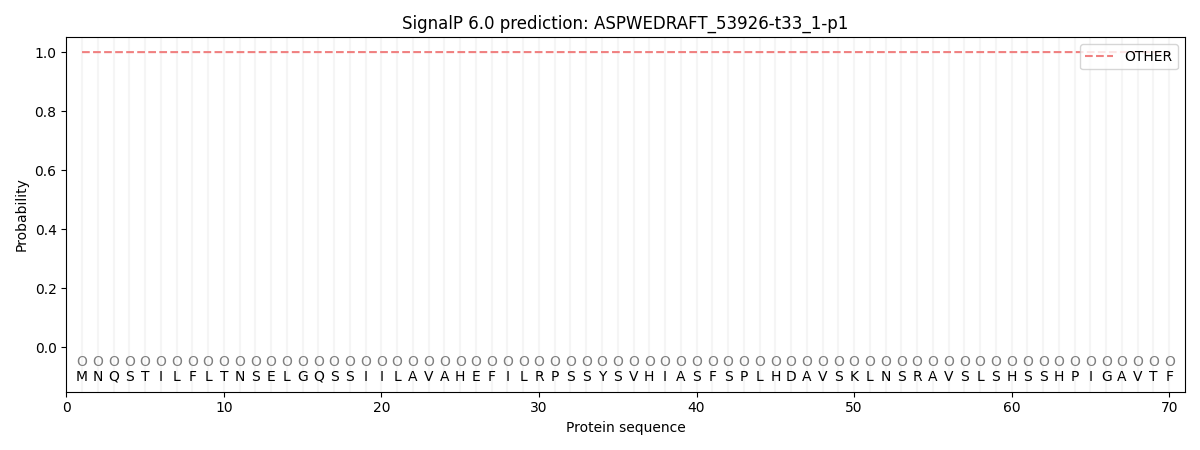
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999947 | 0.000092 |
