You are browsing environment: FUNGIDB
CAZyme Information: ASPWEDRAFT_48851-t33_1-p1
You are here: Home > Sequence: ASPWEDRAFT_48851-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus wentii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus wentii | |||||||||||
| CAZyme ID | ASPWEDRAFT_48851-t33_1-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.149:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 85 | 364 | 8.6e-147 | 0.9964285714285714 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 1.21e-13 | 32 | 371 | 14 | 337 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 4.22e-12 | 78 | 324 | 12 | 243 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.36e-206 | 1 | 389 | 1 | 390 | |
| 3.36e-206 | 1 | 389 | 1 | 390 | |
| 3.36e-206 | 1 | 389 | 1 | 390 | |
| 3.36e-206 | 1 | 389 | 1 | 390 | |
| 3.36e-206 | 1 | 389 | 1 | 390 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.43e-201 | 28 | 389 | 11 | 371 | Chain A, Glycoside hydrolase family 5 [Aspergillus oryzae],6LA0_B Chain B, Glycoside hydrolase family 5 [Aspergillus oryzae] |
|
| 2.43e-185 | 25 | 377 | 11 | 364 | Crystal structure of rutinosidase from Aspergillus niger [Aspergillus niger],6I1A_B Crystal structure of rutinosidase from Aspergillus niger [Aspergillus niger] |
|
| 2.19e-10 | 35 | 319 | 2 | 298 | Chain A, Exo-beta-1,3-glucanase [uncultured bacterium],6ZB9_B Chain B, Exo-beta-1,3-glucanase [uncultured bacterium] |
|
| 1.23e-09 | 35 | 319 | 2 | 298 | Chain A, Exo-beta-1,3-glucanase variant E167Q/E295Q [uncultured bacterium],6ZB8_B Chain B, Exo-beta-1,3-glucanase variant E167Q/E295Q [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.49e-31 | 26 | 318 | 32 | 310 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
|
| 9.35e-26 | 2 | 369 | 8 | 352 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=exgA PE=3 SV=1 |
|
| 1.75e-25 | 2 | 369 | 8 | 352 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
|
| 2.16e-25 | 30 | 374 | 33 | 394 | Glucan 1,3-beta-glucosidase 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=exg1 PE=2 SV=1 |
|
| 2.39e-25 | 17 | 369 | 24 | 352 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
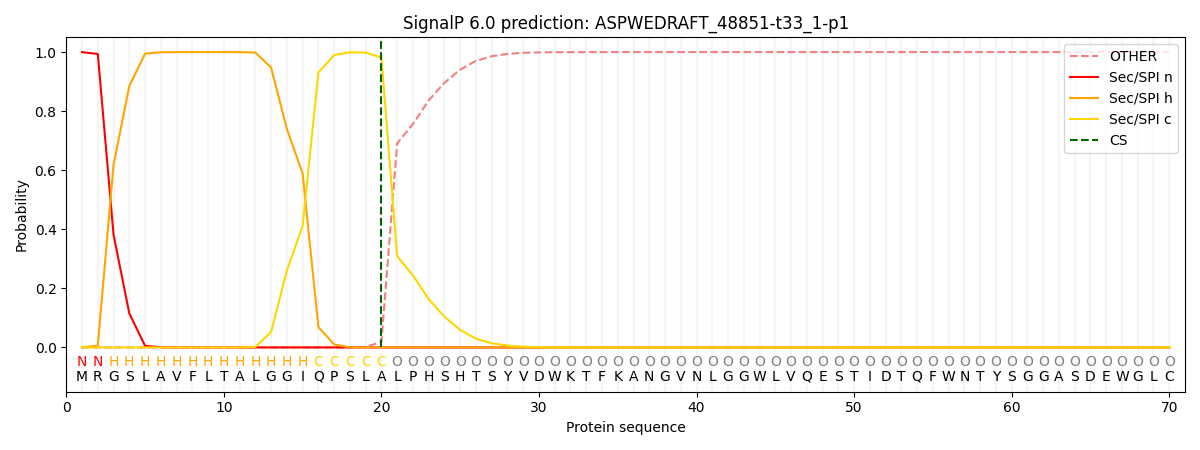
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000258 | 0.999746 | CS pos: 20-21. Pr: 0.9816 |
