You are browsing environment: FUNGIDB
CAZyme Information: ASPWEDRAFT_165629-t33_1-p1
You are here: Home > Sequence: ASPWEDRAFT_165629-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus wentii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus wentii | |||||||||||
| CAZyme ID | ASPWEDRAFT_165629-t33_1-p1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 41 | 335 | 9.6e-81 | 0.9735973597359736 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214750 | Glyco_10 | 1.54e-90 | 80 | 333 | 1 | 262 | Glycosyl hydrolase family 10. |
| 395262 | Glyco_hydro_10 | 1.86e-84 | 37 | 333 | 1 | 307 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 7.67e-73 | 26 | 333 | 13 | 336 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.47e-193 | 7 | 341 | 1 | 331 | |
| 1.62e-193 | 7 | 341 | 1 | 336 | |
| 2.91e-192 | 7 | 341 | 1 | 336 | |
| 2.53e-179 | 7 | 343 | 1 | 338 | |
| 4.41e-172 | 15 | 341 | 6 | 332 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.39e-52 | 37 | 338 | 6 | 320 | Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBR_B Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBU_A Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima],1VBU_B Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima] |
|
| 1.31e-51 | 37 | 338 | 4 | 302 | Crystal Structure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
|
| 1.99e-50 | 37 | 338 | 22 | 336 | Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
|
| 2.25e-49 | 58 | 339 | 26 | 303 | Crystal Structure of xylanase (GH10) in complex with inhibitor (XIP) [Aspergillus nidulans] |
|
| 1.12e-48 | 52 | 333 | 26 | 337 | Highly active enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.94e-49 | 20 | 339 | 4 | 327 | Endo-1,4-beta-xylanase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xlnC PE=1 SV=1 |
|
| 1.73e-45 | 58 | 338 | 49 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
|
| 2.68e-45 | 60 | 338 | 55 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
|
| 5.15e-45 | 58 | 338 | 52 | 326 | Probable endo-1,4-beta-xylanase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xlnC PE=2 SV=2 |
|
| 5.28e-45 | 60 | 338 | 55 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
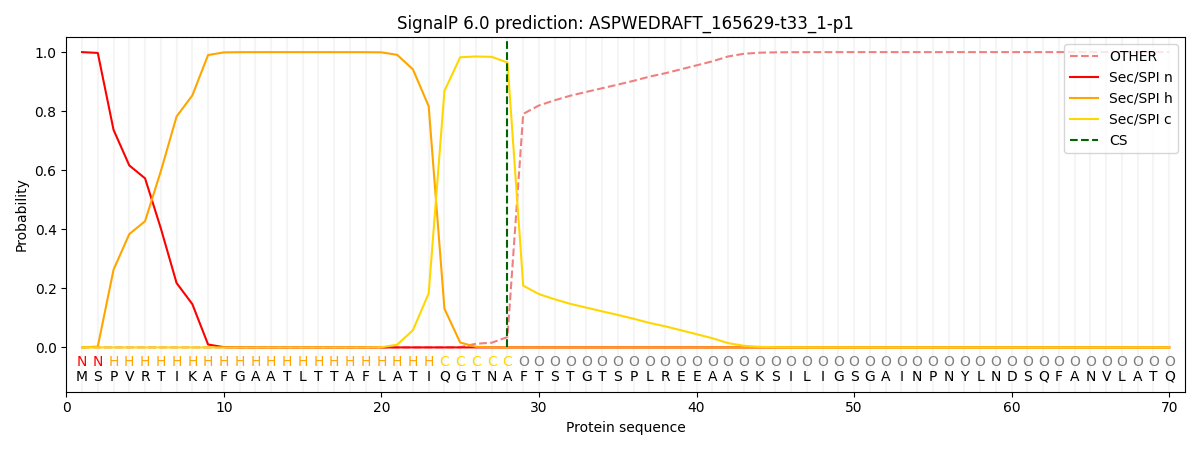
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000459 | 0.999497 | CS pos: 28-29. Pr: 0.9648 |
