You are browsing environment: FUNGIDB
ASPSYDRAFT_718256-t33_1-p1
Basic Information
help
Species
Aspergillus sydowii
Lineage
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus sydowii
CAZyme ID
ASPSYDRAFT_718256-t33_1-p1
CAZy Family
GH95
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
712
80730.36
5.9537
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_AsydowiiCBS593.65
13717
1036612
138
13579
Gene Location
Family
Start
End
Evalue
family coverage
PL27
61
701
9.4e-204
0.9967213114754099
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
271199
LanC_SerThrkinase
7.37e-04
513
669
96
250
Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
6.24e-295
285
710
1
426
1.63e-279
18
707
15
704
1.60e-233
24
704
24
703
5.94e-231
24
704
24
703
5.94e-231
24
704
24
703
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
2.38e-127
13
703
13
690
Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NO8_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838]
more
2.38e-123
13
703
13
690
Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NOK_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838]
more
ASPSYDRAFT_718256-t33_1-p1 has no Swissprot hit.
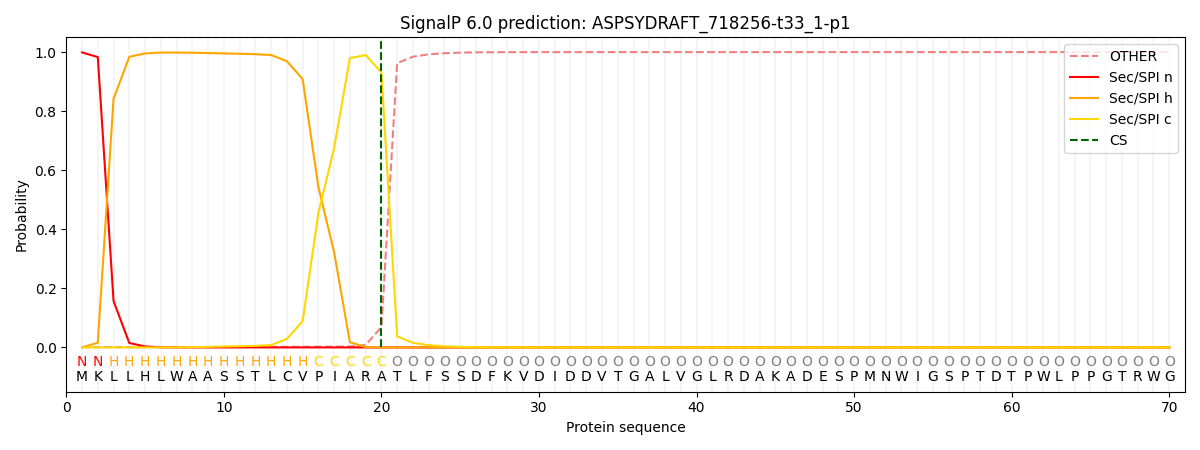
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.002644
0.997318
CS pos: 20-21. Pr: 0.9309
There is no transmembrane helices in ASPSYDRAFT_718256-t33_1-p1.