You are browsing environment: FUNGIDB
CAZyme Information: ASPSYDRAFT_54176-t33_1-p1
You are here: Home > Sequence: ASPSYDRAFT_54176-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus sydowii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus sydowii | |||||||||||
| CAZyme ID | ASPSYDRAFT_54176-t33_1-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.10:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 110 | 298 | 1e-85 | 0.983957219251337 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 4.00e-42 | 111 | 300 | 3 | 189 | Amb_all domain. |
| 226384 | PelB | 9.24e-20 | 104 | 387 | 82 | 344 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 1.49e-16 | 105 | 297 | 15 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.28e-212 | 2 | 387 | 3 | 387 | |
| 1.04e-203 | 8 | 387 | 8 | 389 | |
| 3.70e-202 | 18 | 387 | 20 | 391 | |
| 2.14e-201 | 18 | 387 | 20 | 391 | |
| 2.14e-201 | 18 | 387 | 20 | 391 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.20e-83 | 20 | 387 | 3 | 359 | Pectin Lyase A [Aspergillus niger] |
|
| 2.39e-83 | 18 | 386 | 1 | 359 | Pectin Lyase B [Aspergillus niger] |
|
| 1.34e-82 | 20 | 387 | 3 | 359 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
|
| 9.34e-15 | 111 | 300 | 57 | 249 | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
|
| 1.87e-07 | 113 | 297 | 69 | 276 | Chain A, PECTATE LYASE E [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.11e-224 | 5 | 387 | 6 | 388 | Probable pectin lyase F-1 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelF-1 PE=3 SV=1 |
|
| 4.95e-213 | 1 | 387 | 1 | 386 | Probable pectin lyase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pelF PE=3 SV=1 |
|
| 7.03e-213 | 18 | 387 | 16 | 386 | Probable pectin lyase E OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pelE PE=3 SV=1 |
|
| 3.54e-115 | 20 | 387 | 23 | 380 | Probable pectin lyase F-2 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelF-2 PE=3 SV=1 |
|
| 3.58e-111 | 10 | 387 | 12 | 378 | Probable pectin lyase F OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pelF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
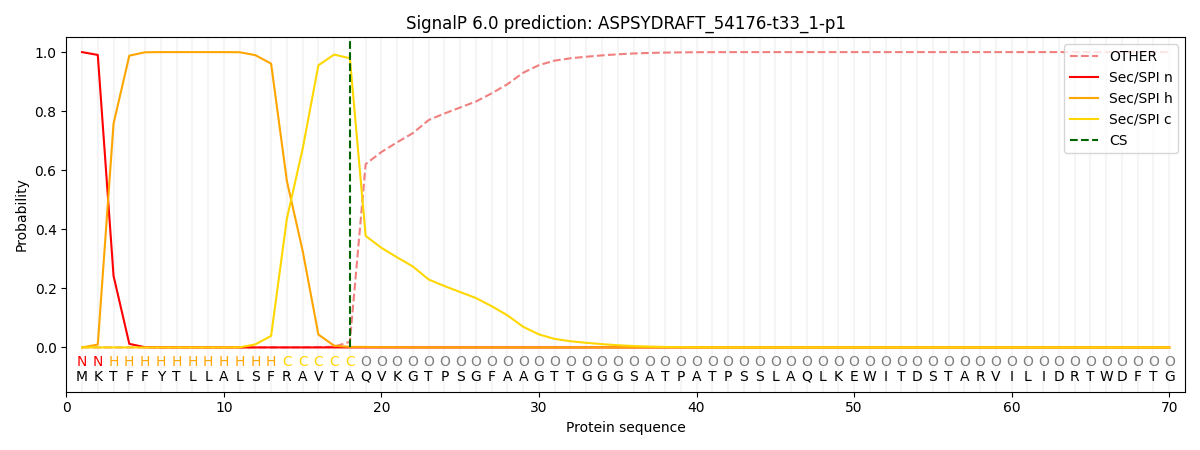
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000262 | 0.999720 | CS pos: 18-19. Pr: 0.9791 |
