You are browsing environment: FUNGIDB
CAZyme Information: ASPSYDRAFT_145302-t33_1-p1
You are here: Home > Sequence: ASPSYDRAFT_145302-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus sydowii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus sydowii | |||||||||||
| CAZyme ID | ASPSYDRAFT_145302-t33_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA16 | 20 | 190 | 6e-73 | 0.9940119760479041 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397269 | LPMO_10 | 3.87e-07 | 20 | 190 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.06e-160 | 1 | 284 | 1 | 283 | |
| 1.54e-111 | 1 | 194 | 1 | 193 | |
| 3.48e-109 | 1 | 280 | 1 | 265 | |
| 1.30e-106 | 1 | 280 | 1 | 285 | |
| 1.30e-106 | 1 | 280 | 1 | 285 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
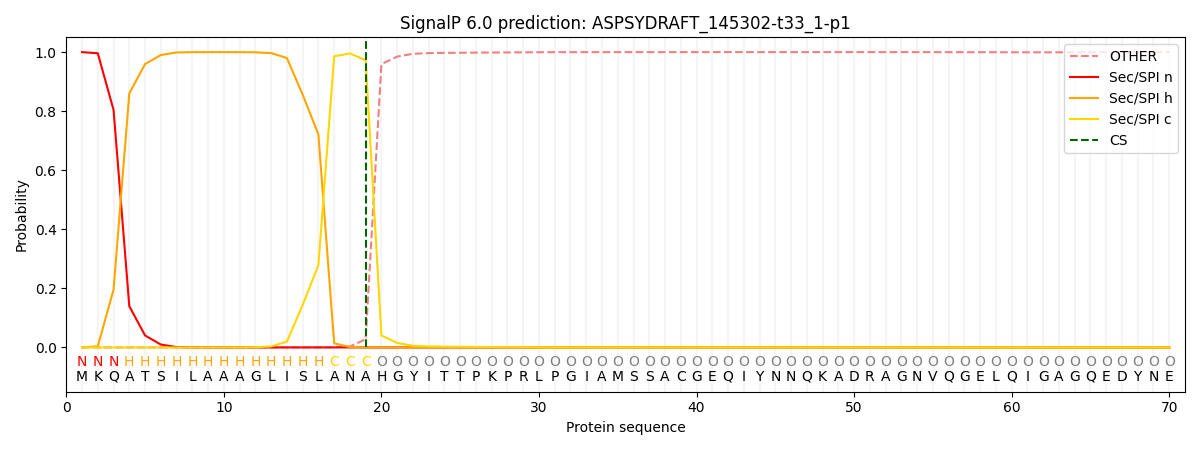
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001069 | 0.998893 | CS pos: 19-20. Pr: 0.9723 |
