You are browsing environment: FUNGIDB
CAZyme Information: ASPSYDRAFT_142918-t33_1-p1
You are here: Home > Sequence: ASPSYDRAFT_142918-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus sydowii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus sydowii | |||||||||||
| CAZyme ID | ASPSYDRAFT_142918-t33_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214773 | CAP10 | 1.58e-14 | 704 | 971 | 30 | 252 | Putative lipopolysaccharide-modifying enzyme. |
| 310354 | Glyco_transf_90 | 1.31e-07 | 706 | 971 | 108 | 322 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
| 340912 | MFS_Mch1p_like | 0.009 | 272 | 412 | 71 | 216 | Monocarboxylate transporter-homologous (Mch) 1 protein and similar transporters of the Major Facilitator Superfamily of transporters. Yeast monocarboxylate transporter-homologous (Mch) proteins are putative transporters that do not transport monocarboxylic acids across the plasma membrane, and may play roles distinct from their mammalian counterparts. Their function has not been determined. The Mch1p-like group belongs to the Monocarboxylate transporter -like (MCT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 984 | 1 | 963 | |
| 0.0 | 1 | 984 | 1 | 873 | |
| 0.0 | 1 | 983 | 1 | 995 | |
| 0.0 | 1 | 983 | 1 | 995 | |
| 0.0 | 5 | 981 | 5 | 991 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.23e-07 | 683 | 970 | 95 | 340 | human POGLUT1 in complex with Notch1 EGF12 and UDP [Homo sapiens],5L0S_A human POGLUT1 in complex with Factor VII EGF1 and UDP [Homo sapiens],5L0T_A human POGLUT1 in complex with EGF(+) and UDP [Homo sapiens],5L0U_A human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose [Homo sapiens],5L0V_A human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP [Homo sapiens],5UB5_A human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP [Homo sapiens] |
|
| 2.63e-06 | 706 | 971 | 154 | 378 | Crystal structure of Drosophila Poglut1 (Rumi) complexed with its glycoprotein product (glucosylated EGF repeat) and UDP [Drosophila melanogaster],5F85_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) and UDP [Drosophila melanogaster],5F86_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) [Drosophila melanogaster],5F87_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_B Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_C Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_D Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_E Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_F Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.29e-36 | 421 | 977 | 151 | 661 | Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
|
| 1.28e-09 | 701 | 971 | 158 | 385 | O-glucosyltransferase rumi homolog OS=Anopheles gambiae OX=7165 GN=AGAP004267 PE=3 SV=1 |
|
| 5.33e-09 | 675 | 972 | 131 | 388 | O-glucosyltransferase rumi homolog OS=Aedes aegypti OX=7159 GN=AAEL011121 PE=3 SV=1 |
|
| 7.08e-09 | 838 | 972 | 264 | 389 | O-glucosyltransferase rumi homolog OS=Culex quinquefasciatus OX=7176 GN=CPIJ013394 PE=3 SV=1 |
|
| 1.96e-06 | 844 | 971 | 272 | 388 | O-glucosyltransferase rumi OS=Drosophila pseudoobscura pseudoobscura OX=46245 GN=rumi PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
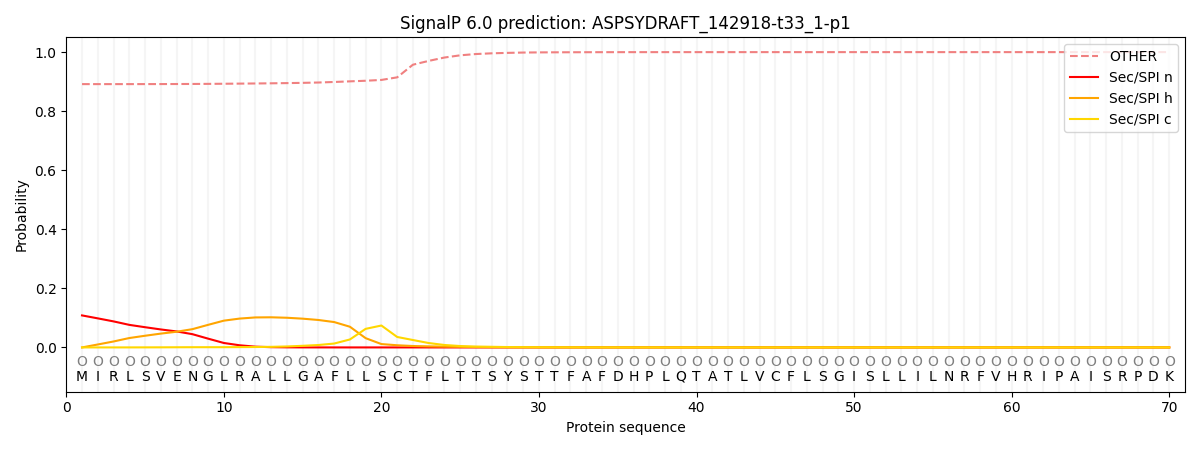
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.891972 | 0.108013 |
TMHMM Annotations download full data without filtering help
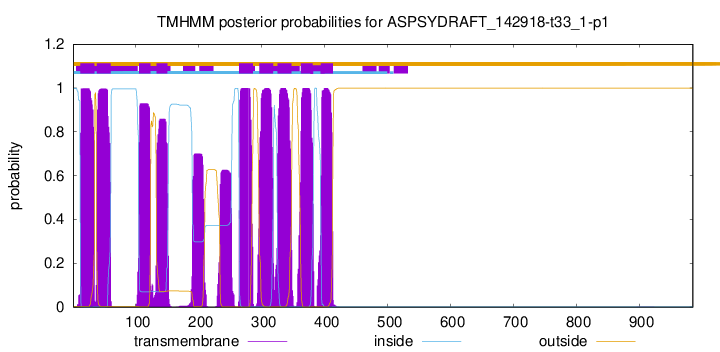
| Start | End |
|---|---|
| 12 | 34 |
| 38 | 60 |
| 105 | 123 |
| 133 | 150 |
| 264 | 286 |
| 296 | 318 |
| 325 | 347 |
| 362 | 381 |
| 394 | 413 |
