You are browsing environment: FUNGIDB
CAZyme Information: ASPSYDRAFT_139049-t33_1-p1
You are here: Home > Sequence: ASPSYDRAFT_139049-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus sydowii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus sydowii | |||||||||||
| CAZyme ID | ASPSYDRAFT_139049-t33_1-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 124 | 569 | 1.7e-56 | 0.9454148471615721 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396238 | FAD_binding_4 | 7.61e-19 | 125 | 267 | 7 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 369658 | BBE | 1.62e-09 | 531 | 568 | 1 | 38 | Berberine and berberine like. This domain is found in the berberine bridge and berberine bridge- like enzymes which are involved in the biosynthesis of numerous isoquinoline alkaloids. They catalyze the transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine. |
| 223354 | GlcD | 1.94e-09 | 126 | 266 | 39 | 169 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 215242 | PLN02441 | 2.67e-04 | 126 | 277 | 72 | 220 | cytokinin dehydrogenase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.18e-58 | 13 | 580 | 492 | 1055 | |
| 5.18e-58 | 13 | 580 | 492 | 1055 | |
| 1.56e-17 | 125 | 576 | 76 | 497 | |
| 3.11e-13 | 126 | 568 | 171 | 586 | |
| 1.74e-12 | 94 | 574 | 44 | 488 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.09e-81 | 19 | 586 | 25 | 573 | Crystal structure of VAO-type flavoprotein MtVAO615 at pH 7.5 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_A Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F73_B Crystal structure of VAO-type flavoprotein MtVAO615 at pH 5.0 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
|
| 1.51e-57 | 22 | 579 | 30 | 587 | Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_B Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_C Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],6F74_D Crystal structure of VAO-type flavoprotein MtVAO713 from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464] |
|
| 2.41e-16 | 128 | 303 | 87 | 253 | Structure of BBE-like #28 from Arabidopsis thaliana [Arabidopsis thaliana],5D79_B Structure of BBE-like #28 from Arabidopsis thaliana [Arabidopsis thaliana] |
|
| 3.51e-16 | 126 | 568 | 55 | 462 | Physcomitrella patens BBE-like 1 variant D396N [Physcomitrium patens],6EO5_B Physcomitrella patens BBE-like 1 variant D396N [Physcomitrium patens] |
|
| 3.81e-16 | 121 | 568 | 33 | 491 | Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6],5I1V_B Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6],5I1V_C Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6],5I1V_D Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6],5I1W_A Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6],5I1W_B Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6],5I1W_C Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6],5I1W_D Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis [Actinoalloteichus sp. WH1-2216-6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.44e-196 | 22 | 586 | 26 | 587 | Uncharacterized FAD-linked oxidoreductase ARB_02478 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07056 PE=1 SV=2 |
|
| 7.11e-142 | 19 | 585 | 22 | 576 | FAD-linked oxidoreductase malF OS=Malbranchea aurantiaca OX=78605 GN=malF PE=1 SV=1 |
|
| 2.91e-124 | 12 | 585 | 18 | 587 | FAD-linked oxidoreductase notD OS=Aspergillus sp. (strain MF297-2) OX=877550 GN=notD PE=1 SV=1 |
|
| 5.86e-123 | 22 | 585 | 27 | 581 | FAD-linked oxidoreductase easE OS=Epichloe festucae var. lolii OX=73839 GN=easE PE=2 SV=1 |
|
| 6.01e-118 | 4 | 586 | 7 | 580 | FAD-linked oxidoreductase notD' OS=Aspergillus versicolor OX=46472 GN=notD' PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
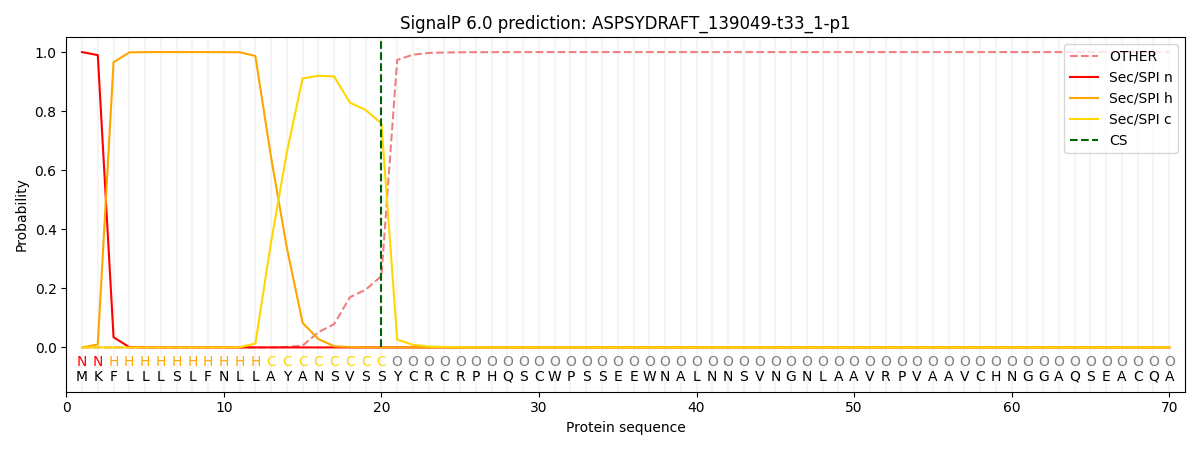
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000239 | 0.999759 | CS pos: 20-21. Pr: 0.7582 |
