You are browsing environment: FUNGIDB
CAZyme Information: ASPNIDRAFT2_1163981-t41_1-p1
You are here: Home > Sequence: ASPNIDRAFT2_1163981-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
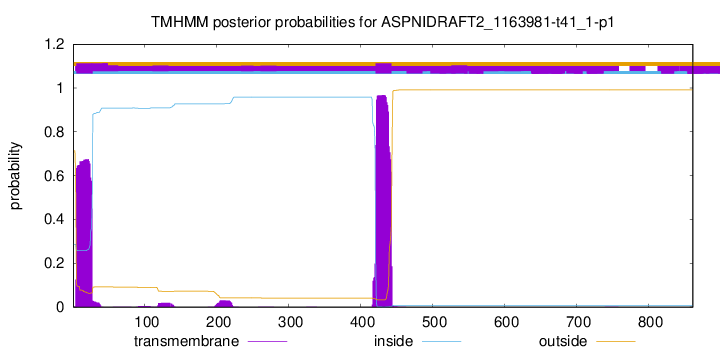
TMHMM annotations
Basic Information help
| Species | Aspergillus niger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger | |||||||||||
| CAZyme ID | ASPNIDRAFT2_1163981-t41_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | Glycosyltransferase Family 62 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 461 | 724 | 1.2e-124 | 0.9888059701492538 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397491 | Anp1 | 1.87e-158 | 462 | 724 | 1 | 265 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 212492 | retinol-DH_like_SDR_c_like | 1.02e-27 | 8 | 261 | 3 | 220 | retinol dehydrogenase (retinol-DH), Light dependent Protochlorophyllide (Pchlide) OxidoReductase (LPOR) and related proteins, classical (c) SDRs. Classical SDR subgroup containing retinol-DHs, LPORs, and related proteins. Retinol is processed by a medium chain alcohol dehydrogenase followed by retinol-DHs. Pchlide reductases act in chlorophyll biosynthesis. There are distinct enzymes that catalyze Pchlide reduction in light or dark conditions. Light-dependent reduction is via an NADP-dependent SDR, LPOR. Proteins in this subfamily share the glycine-rich NAD-binding motif of the classical SDRs, have a partial match to the canonical active site tetrad, but lack the typical active site Ser. This subgroup includes the human proteins: retinol dehydrogenase -12, -13 ,and -14, dehydrogenase/reductase SDR family member (DHRS)-12 , -13 and -X (a DHRS on chromosome X), and WWOX (WW domain-containing oxidoreductase), as well as a Neurospora crassa SDR encoded by the blue light inducible bli-4 gene. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold (alpha/beta folding pattern with a central beta-sheet), an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Classical SDRs are typically about 250 residues long, while extended SDRs are approximately 350 residues. Sequence identity between different SDR enzymes are typically in the 15-30% range, but the enzymes share the Rossmann fold NAD-binding motif and characteristic NAD-binding and catalytic sequence patterns. These enzymes catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human 15-hydroxyprostaglandin dehydrogenase (15-PGDH) numbering). In addition to the Tyr and Lys, there is often an upstream Ser (Ser-138, 15-PGDH numbering) and/or an Asn (Asn-107, 15-PGDH numbering) contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. Extended SDRs have additional elements in the C-terminal region, and typically have a TGXXGXXG cofactor binding motif. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. Some atypical SDRs have lost catalytic activity and/or have an unusual NAD(P)-binding motif and missing or unusual active site residues. Reactions catalyzed within the SDR family include isomerization, decarboxylation, epimerization, C=N bond reduction, dehydratase activity, dehalogenation, Enoyl-CoA reduction, and carbonyl-alcohol oxidoreduction. |
| 235506 | fabG | 1.72e-12 | 8 | 167 | 7 | 165 | 3-ketoacyl-(acyl-carrier-protein) reductase; Provisional |
| 223377 | DltE | 1.36e-11 | 1 | 155 | 1 | 142 | Short-chain dehydrogenase [General function prediction only]. |
| 212491 | SDR_c | 1.89e-11 | 9 | 160 | 1 | 135 | classical (c) SDRs. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold (alpha/beta folding pattern with a central beta-sheet), an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Classical SDRs are typically about 250 residues long, while extended SDRs are approximately 350 residues. Sequence identity between different SDR enzymes are typically in the 15-30% range, but the enzymes share the Rossmann fold NAD-binding motif and characteristic NAD-binding and catalytic sequence patterns. These enzymes catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human prostaglandin dehydrogenase (PGDH) numbering). In addition to the Tyr and Lys, there is often an upstream Ser (Ser-138, PGDH numbering) and/or an Asn (Asn-107, PGDH numbering) contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. Extended SDRs have additional elements in the C-terminal region, and typically have a TGXXGXXG cofactor binding motif. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. Some atypical SDRs have lost catalytic activity and/or have an unusual NAD(P)-binding motif and missing or unusual active site residues. Reactions catalyzed within the SDR family include isomerization, decarboxylation, epimerization, C=N bond reduction, dehydratase activity, dehalogenation, Enoyl-CoA reduction, and carbonyl-alcohol oxidoreduction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 816 | 1 | 737 | |
| 0.0 | 372 | 862 | 1 | 491 | |
| 8.97e-314 | 372 | 862 | 1 | 496 | |
| 8.97e-314 | 372 | 862 | 1 | 496 | |
| 5.16e-253 | 372 | 833 | 1 | 459 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.73e-58 | 463 | 760 | 6 | 304 | Crystal structure of Saccharomyces cerevisiae Mnn9 in complex with GDP and Mn. [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.77e-154 | 462 | 814 | 61 | 413 | Mannan polymerase II complex ANP1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ANP1 PE=1 SV=3 |
|
| 9.69e-151 | 396 | 767 | 14 | 383 | Mannan polymerase II complex anp1 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=anp1 PE=3 SV=1 |
|
| 7.41e-117 | 462 | 767 | 171 | 523 | Mannan polymerase I complex VAN1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=VAN1 PE=1 SV=3 |
|
| 1.43e-114 | 454 | 767 | 67 | 430 | Vanadate resistance protein OS=Candida albicans OX=5476 GN=VAN1 PE=3 SV=1 |
|
| 1.60e-63 | 456 | 758 | 66 | 368 | Mannan polymerase complex subunit MNN9 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN9 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
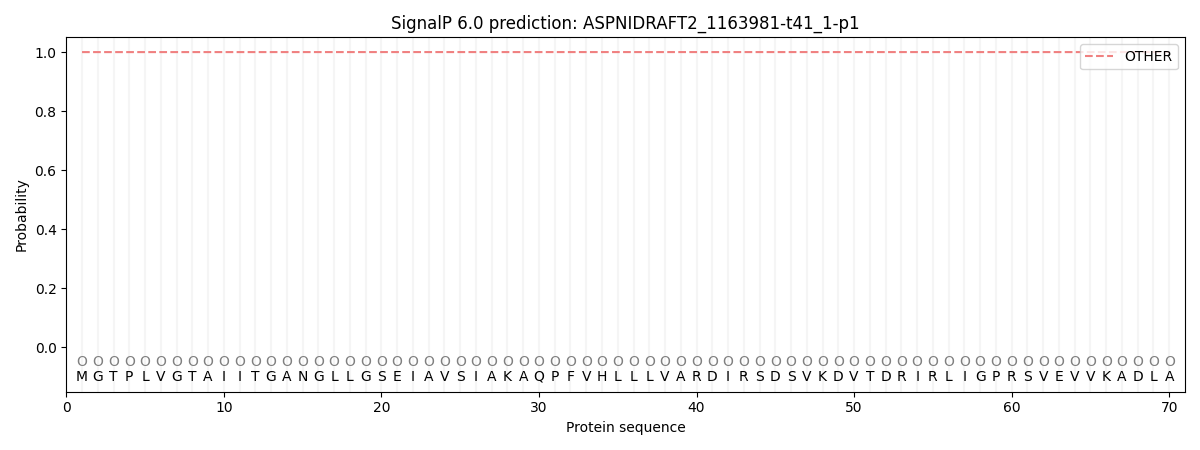
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000066 | 0.000000 |