You are browsing environment: FUNGIDB
CAZyme Information: ASPGLDRAFT_21213-t33_1-p1
You are here: Home > Sequence: ASPGLDRAFT_21213-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus glaucus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus glaucus | |||||||||||
| CAZyme ID | ASPGLDRAFT_21213-t33_1-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.72:1 | 3.2.1.8:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 66 | 200 | 1.3e-19 | 0.5991189427312775 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 5.12e-35 | 65 | 328 | 43 | 306 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 223477 | YpfH | 6.65e-05 | 84 | 240 | 17 | 167 | Predicted esterase [General function prediction only]. |
| 225256 | Fes | 0.002 | 66 | 169 | 79 | 189 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.52e-27 | 62 | 331 | 28 | 291 | |
| 2.03e-20 | 62 | 327 | 181 | 449 | |
| 5.23e-20 | 34 | 331 | 164 | 460 | |
| 5.23e-20 | 34 | 331 | 164 | 460 | |
| 5.23e-20 | 34 | 331 | 164 | 460 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.66e-08 | 66 | 238 | 18 | 195 | C-terminal esterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.67e-17 | 69 | 300 | 292 | 519 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
|
| 2.35e-13 | 64 | 305 | 53 | 269 | Bifunctional acetylxylan esterase/xylanase XynS20E OS=Neocallimastix patriciarum OX=4758 GN=xynS20E PE=1 SV=1 |
|
| 5.58e-12 | 47 | 238 | 21 | 200 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
|
| 1.84e-11 | 68 | 268 | 42 | 228 | Probable feruloyl esterase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=faeC PE=3 SV=1 |
|
| 1.88e-11 | 37 | 238 | 23 | 200 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
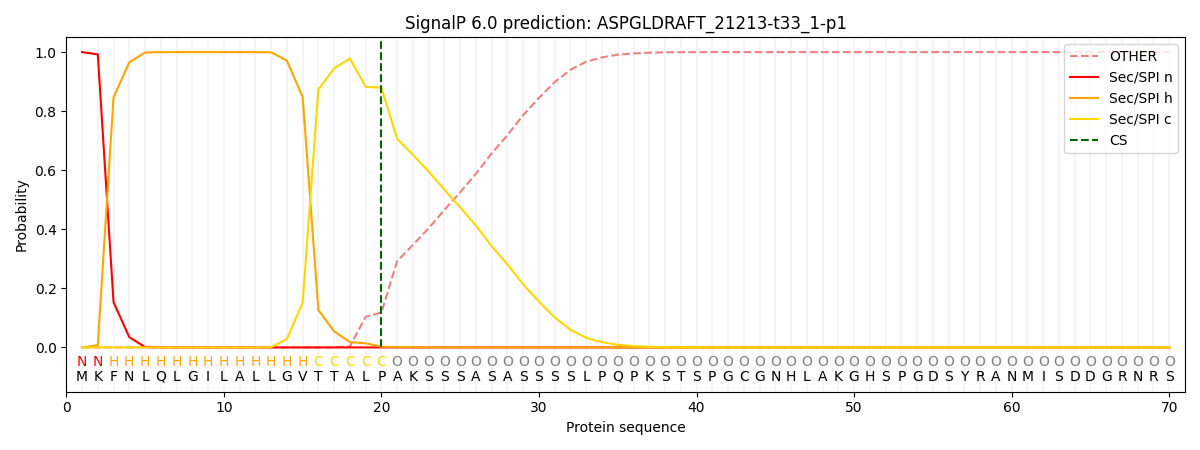
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000214 | 0.999743 | CS pos: 20-21. Pr: 0.8792 |
