You are browsing environment: FUNGIDB
CAZyme Information: ASPGLDRAFT_134307-t33_1-p1
You are here: Home > Sequence: ASPGLDRAFT_134307-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus glaucus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus glaucus | |||||||||||
| CAZyme ID | ASPGLDRAFT_134307-t33_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 28 | 214 | 3e-79 | 0.9896907216494846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 9.08e-114 | 21 | 219 | 1 | 200 | Pectate lyase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.71e-167 | 1 | 241 | 1 | 252 | |
| 1.56e-125 | 1 | 241 | 1 | 242 | |
| 1.56e-125 | 1 | 241 | 1 | 242 | |
| 1.56e-125 | 1 | 241 | 1 | 242 | |
| 1.56e-125 | 1 | 241 | 1 | 242 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.10e-22 | 42 | 161 | 16 | 135 | Crystal Structure Of Pectate Lyase From Bacillus Sp. Strain Ksm-P15. [Bacillus sp. KSM-P15] |
|
| 2.14e-21 | 52 | 225 | 12 | 208 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B90_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
|
| 1.31e-20 | 52 | 225 | 130 | 326 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B4N_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_A Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
|
| 6.14e-20 | 39 | 162 | 10 | 139 | The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725],4EW9_B The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725] |
|
| 6.27e-20 | 39 | 162 | 11 | 140 | The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii],3T9G_B The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.59e-132 | 1 | 241 | 1 | 242 | Probable pectate lyase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyD PE=3 SV=1 |
|
| 1.59e-132 | 1 | 241 | 1 | 242 | Probable pectate lyase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyD PE=3 SV=1 |
|
| 1.26e-131 | 1 | 241 | 1 | 241 | Probable pectate lyase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyD PE=3 SV=1 |
|
| 4.29e-128 | 1 | 241 | 1 | 253 | Probable pectate lyase D OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyD PE=3 SV=1 |
|
| 7.05e-124 | 12 | 241 | 28 | 260 | Pectate lyase H OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
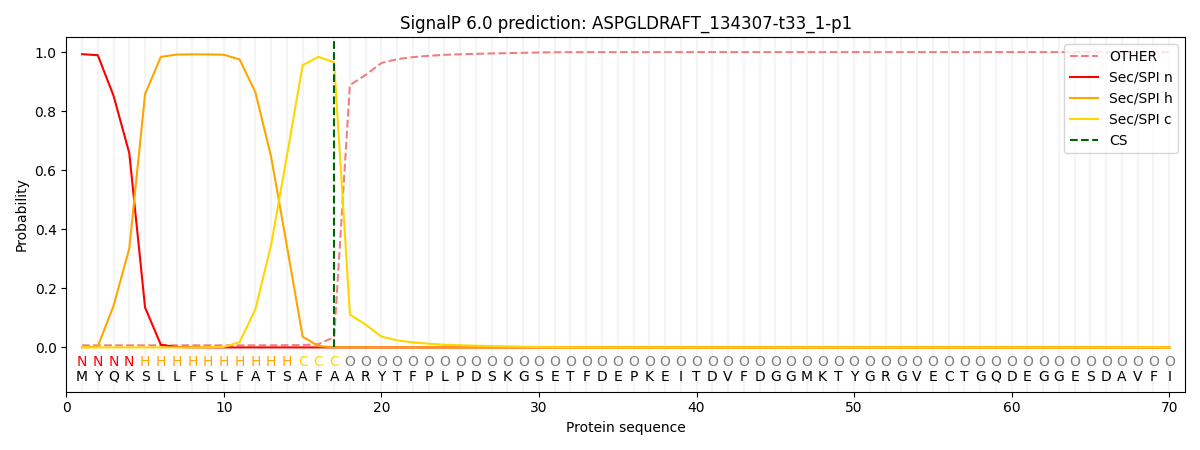
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.013090 | 0.986892 | CS pos: 17-18. Pr: 0.9651 |
