You are browsing environment: FUNGIDB
CAZyme Information: ASPFODRAFT_127559-t33_1-p1
Basic Information
help
| Species |
Aspergillus luchuensis
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus luchuensis
|
| CAZyme ID |
ASPFODRAFT_127559-t33_1-p1
|
| CAZy Family |
AA3 |
| CAZyme Description |
glycoside hydrolase family 5 protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 412 |
|
45810.75 |
4.0169 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AluchuensisCBS106.47 |
13784 |
1137211 |
282 |
13502
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
33 |
367 |
1.6e-168 |
0.9970326409495549 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 395098
|
Cellulase |
5.15e-05 |
34 |
364 |
1 |
269 |
Cellulase (glycosyl hydrolase family 5). |
| 225344
|
BglC |
0.001 |
34 |
382 |
38 |
378 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.54e-309 |
1 |
412 |
1 |
412 |
| 1.54e-309 |
1 |
412 |
1 |
412 |
| 8.07e-304 |
1 |
412 |
1 |
412 |
| 5.75e-220 |
12 |
412 |
12 |
412 |
| 2.02e-215 |
12 |
412 |
12 |
430 |
ASPFODRAFT_127559-t33_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 9.02e-50 |
1 |
399 |
11 |
388 |
Glycosyl hydrolase 5 family protein OS=Chamaecyparis obtusa OX=13415 PE=1 SV=1 |
| 1.93e-06 |
26 |
254 |
630 |
840 |
Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
This protein is predicted as SP
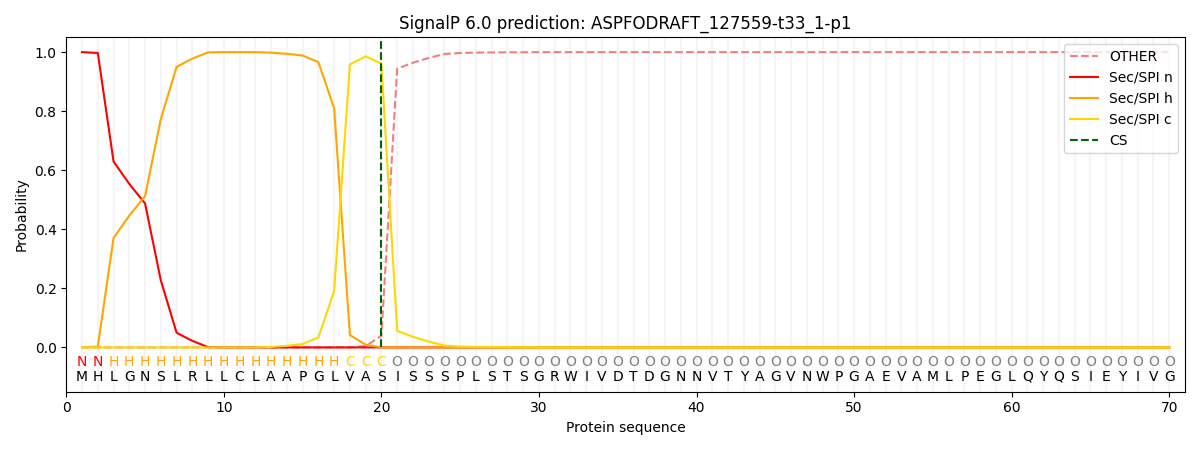
| Other |
SP_Sec_SPI |
CS Position |
| 0.000283 |
0.999672 |
CS pos: 20-21. Pr: 0.9605 |
There is no transmembrane helices in ASPFODRAFT_127559-t33_1-p1.

