You are browsing environment: FUNGIDB
CAZyme Information: ASPCADRAFT_212219-t33_1-p1
Basic Information
help
| Species |
Aspergillus carbonarius
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus carbonarius
|
| CAZyme ID |
ASPCADRAFT_212219-t33_1-p1
|
| CAZy Family |
GH54|CBM42 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 678 |
|
75274.79 |
4.2596 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AcarbonariusITEM5010 |
11735 |
602072 |
260 |
11475
|
|
| Gene Location |
No EC number prediction in ASPCADRAFT_212219-t33_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH142 |
28 |
510 |
8.8e-154 |
0.964509394572025 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 225942
|
GDB1 |
0.001 |
178 |
420 |
372 |
588 |
Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
677 |
1 |
679 |
| 0.0 |
1 |
677 |
1 |
679 |
| 0.0 |
12 |
677 |
14 |
678 |
| 0.0 |
12 |
677 |
31 |
695 |
| 0.0 |
12 |
677 |
14 |
678 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 3.05e-67 |
33 |
511 |
653 |
1101 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
| 1.20e-63 |
33 |
511 |
653 |
1101 |
Sialidase BT_1020 [Bacteroides thetaiotaomicron] |
ASPCADRAFT_212219-t33_1-p1 has no Swissprot hit.
This protein is predicted as SP
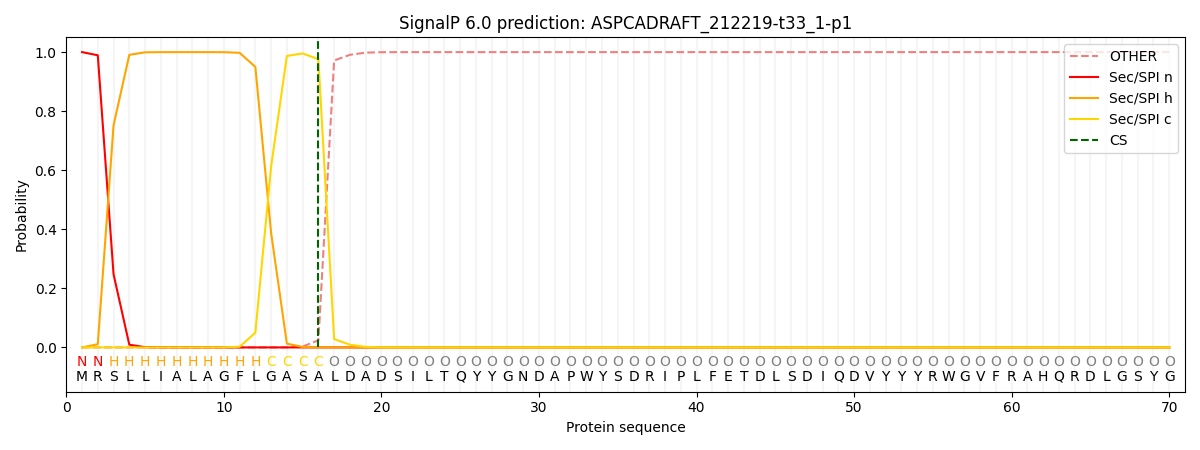
| Other |
SP_Sec_SPI |
CS Position |
| 0.000263 |
0.999696 |
CS pos: 16-17. Pr: 0.9758 |
There is no transmembrane helices in ASPCADRAFT_212219-t33_1-p1.

