You are browsing environment: FUNGIDB
CAZyme Information: ASPCADRAFT_212173-t33_1-p1
You are here: Home > Sequence: ASPCADRAFT_212173-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
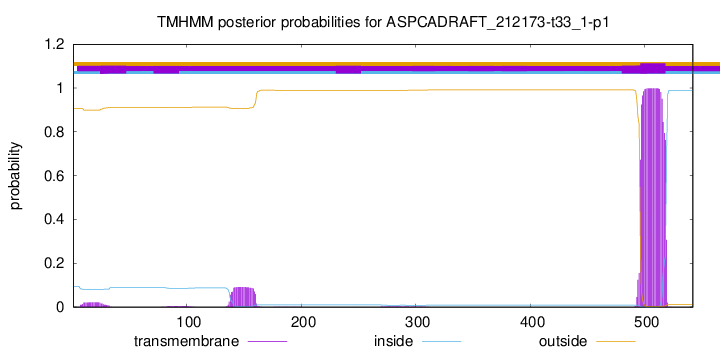
TMHMM annotations
Basic Information help
| Species | Aspergillus carbonarius | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus carbonarius | |||||||||||
| CAZyme ID | ASPCADRAFT_212173-t33_1-p1 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | glycosyltransferase family 1 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 84 | 433 | 2.3e-22 | 0.7774869109947644 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 3.35e-38 | 10 | 459 | 4 | 403 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 278624 | UDPGT | 6.51e-12 | 336 | 476 | 312 | 451 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 224732 | YjiC | 4.38e-11 | 7 | 461 | 3 | 399 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 215465 | PLN02863 | 6.88e-09 | 244 | 399 | 245 | 401 | UDP-glucoronosyl/UDP-glucosyl transferase family protein |
| 178572 | PLN02992 | 1.37e-08 | 240 | 438 | 239 | 438 | coniferyl-alcohol glucosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.34e-262 | 3 | 522 | 7 | 530 | |
| 2.83e-249 | 1 | 524 | 1 | 525 | |
| 2.40e-237 | 3 | 525 | 6 | 533 | |
| 6.04e-236 | 3 | 522 | 7 | 530 | |
| 2.04e-232 | 4 | 522 | 9 | 511 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.39e-11 | 2 | 447 | 12 | 454 | Chain A, indoxyl UDP-glucosyltransferase [Persicaria tinctoria],5NLM_B Chain B, indoxyl UDP-glucosyltransferase [Persicaria tinctoria] |
|
| 4.49e-11 | 9 | 447 | 38 | 475 | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and UDP-glucose [Persicaria tinctoria],6SU6_B Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and UDP-glucose [Persicaria tinctoria],6SU7_A Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline [Persicaria tinctoria],6SU7_B Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline [Persicaria tinctoria],6SU7_C Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline [Persicaria tinctoria],6SU7_D Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline [Persicaria tinctoria] |
|
| 2.50e-07 | 268 | 399 | 273 | 389 | Chain A, Glycosyltransferase [Oryza sativa Japonica Group],7ERY_A Chain A, Glycosyltransferase [Oryza sativa Japonica Group],7ES0_A Chain A, Glycosyltransferase [Oryza sativa Japonica Group],7ES1_A Chain A, Glycosyltransferase [Oryza sativa Japonica Group] |
|
| 2.50e-07 | 268 | 399 | 273 | 389 | Chain A, Glycosyltransferase [Oryza sativa Japonica Group] |
|
| 1.85e-06 | 222 | 394 | 237 | 418 | Structure of Phytolacca americana UGT3 with 15-crown-5 [Phytolacca americana],6LZX_B Structure of Phytolacca americana UGT3 with 15-crown-5 [Phytolacca americana],6LZY_A Structure of Phytolacca americana UGT3 with 18-crown-6 [Phytolacca americana],6LZY_B Structure of Phytolacca americana UGT3 with 18-crown-6 [Phytolacca americana],7VEJ_A Chain A, Glycosyltransferase [Phytolacca americana],7VEJ_B Chain B, Glycosyltransferase [Phytolacca americana],7VEK_A Chain A, Glycosyltransferase [Phytolacca americana],7VEK_B Chain B, Glycosyltransferase [Phytolacca americana],7VEL_A Chain A, Glycosyltransferase [Phytolacca americana],7VEL_B Chain B, Glycosyltransferase [Phytolacca americana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.02e-09 | 154 | 438 | 188 | 441 | UDP-glucuronosyltransferase 1-6 OS=Oryctolagus cuniculus OX=9986 GN=UGT1 PE=1 SV=1 |
|
| 5.28e-09 | 334 | 505 | 329 | 489 | UDP-glycosyltransferase UGT5 OS=Dactylopius coccus OX=765876 GN=UGT5 PE=1 SV=1 |
|
| 1.62e-08 | 151 | 442 | 187 | 448 | UDP-glucuronosyltransferase 2B4 OS=Homo sapiens OX=9606 GN=UGT2B4 PE=1 SV=2 |
|
| 3.41e-08 | 232 | 458 | 235 | 458 | UDP-glycosyltransferase 91D2 OS=Stevia rebaudiana OX=55670 GN=UGT91D2 PE=1 SV=1 |
|
| 3.73e-08 | 350 | 442 | 356 | 446 | UDP-glucuronosyltransferase 2A3 OS=Homo sapiens OX=9606 GN=UGT2A3 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999694 | 0.000352 |