You are browsing environment: FUNGIDB
CAZyme Information: ASPBRDRAFT_52061-t33_1-p1
You are here: Home > Sequence: ASPBRDRAFT_52061-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus brasiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus brasiliensis | |||||||||||
| CAZyme ID | ASPBRDRAFT_52061-t33_1-p1 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT59 | 34 | 537 | 3e-146 | 0.995049504950495 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 399229 | Tyr-DNA_phospho | 0.0 | 704 | 1164 | 1 | 433 | Tyrosyl-DNA phosphodiesterase. Covalent intermediates between topoisomerase I and DNA can become dead-end complexes that lead to cell death. Tyrosyl-DNA phosphodiesterase can hydrolyze the bond between topoisomerase I and DNA. |
| 398537 | DIE2_ALG10 | 1.35e-138 | 35 | 537 | 1 | 383 | DIE2/ALG10 family. The ALG10 protein from Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum. This protein has been characterized in rat as potassium channel regulator 1. |
| 197290 | PLDc_yTdp1_1 | 5.78e-58 | 703 | 877 | 1 | 166 | Catalytic domain, repeat 1, of yeast tyrosyl-DNA phosphodiesterase. Catalytic domain, repeat 1, of yeast tyrosyl-DNA phosphodiesterase (yTdp1, EC 3.1.4.-). yTdp1 is involved in the repair of topoisomerase I DNA lesions by hydrolyzing the topoisomerase from the 3'-end of the DNA during double-strand break repair. Unlike human Tdp1 whose substrate-binding pocket can accommodate a fairly large topoisomerase I peptide fragment, yTdp1 has a preference for substrates containing one to four amino acid residues. The monomeric yTdp1 contains two copies of a variant HKD motif (H-x-K-x(4)-D, where x represents any amino acid residue), which consists of the highly conserved histidine and lysine residues, but lacks the aspartate residue that is well conserved in other phospholipase D (PLD, EC 3.1.4.4) enzymes. Like other PLD enzymes, yTdp1 may utilize a common two-step general acid/base catalytic mechanism, involving a DNA-enzyme intermediate to cleave phosphodiester bonds. A single active site involved in phosphatidyl group transfer would be formed by the two variant HKD motifs from the N- and C-terminal domains in a pseudodimeric way. |
| 197222 | PLDc_Tdp1_2 | 4.77e-49 | 926 | 1104 | 1 | 182 | Catalytic domain, repeat 2, of tyrosyl-DNA phosphodiesterase. Catalytic domain, repeat 2, of Tyrosyl-DNA phosphodiesterase (Tdp1, EC 3.1.4.-), which exists in eukaryotes but not in prokaryotes. Tdp1 acts as an important DNA repair enzyme that removes stalled topoisomerase I-DNA complexes by catalyzing the hydrolysis of a phosphodiester bond between a tyrosine side chain and a DNA 3'-phosphate. It is a monomeric protein that contains two copies of a variant HKD motif (H-x-K-x(4)-D, where x represents any amino acid residue), which consists of the highly conserved histidine and lysine residues, but lacks the aspartate residue that is well conserved in other phospholipase D (PLD, EC 3.1.4.4) enzymes. Thus, this family represents a distinct class within the PLD superfamily. Like other PLD enzymes, Tdp1 may utilize a common two-step general acid/base catalytic mechanism, involving a DNA-enzyme intermediate to cleave phosphodiester bonds. A single active site involved in phosphatidyl group transfer would be formed by the two variant HKD motifs from the N- and C-terminal domains in a pseudodimeric way. |
| 197291 | PLDc_mTdp1_2 | 1.72e-39 | 902 | 1105 | 3 | 191 | Catalytic domain, repeat 2, of metazoan tyrosyl-DNA phosphodiesterase. Catalytic domain, repeat 2, of metazoan tyrosyl-DNA phosphodiesterase (Tdp1, EC 3.1.4.-). Human Tdp1 (hTdp1) acts as an important DNA repair enzyme with a preference for single-stranded or blunt-ended duplex oligonucleotides. It can remove stalled topoisomerase I-DNA complexes by catalyzing the hydrolysis of a phosphodiester bond between a tyrosine side chain and a DNA 3'-phosphate. It is therefore a potential molecular target for new anti-cancer drugs. hTdp1 has been shown to associate with additional proteins, such as XRCC1, to form a multi-enzyme complex. These additional proteins may be involved in recognizing 3'-phoshotyrosyl DNA in vivo. hTdp1 is a monomeric protein containing two copies of a variant HKD motif (H-x-K-x(4)-D, where x represents any amino acid residue), which consists of the highly conserved histidine and lysine residues, but lacks the aspartate residue that is well conserved in other phospholipase D (PLD, EC 3.1.4.4) enzymes. Like other PLD enzymes, hTdp1 may utilize a common two-step general acid/base catalytic mechanism, involving a DNA-enzyme intermediate to cleave phosphodiester bonds. A single active site involved in phosphatidyl group transfer would be formed by the two variant HKD motifs from the N- and C-terminal domains in a pseudodimeric way. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 605 | 1 | 607 | |
| 0.0 | 1 | 605 | 1 | 607 | |
| 0.0 | 1 | 605 | 1 | 565 | |
| 1.69e-312 | 3 | 605 | 4 | 607 | |
| 2.39e-312 | 3 | 605 | 4 | 607 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.12e-56 | 703 | 1189 | 14 | 457 | Crystal structure of Tdp1 catalytic domain in complex with compound XZ519 [Homo sapiens],6MJ5_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ519 [Homo sapiens] |
|
| 1.15e-56 | 703 | 1189 | 15 | 458 | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT0911 from cocktail soak [Homo sapiens],6DHU_B Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT0911 from cocktail soak [Homo sapiens],6DIE_A Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment benzene-1,2,4-tricarboxylic acid from single soak [Homo sapiens],6DIE_B Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment benzene-1,2,4-tricarboxylic acid from single soak [Homo sapiens],6DIH_A Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound PH004941 [Homo sapiens],6DIH_B Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound PH004941 [Homo sapiens],6DIM_A Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT1982 from cocktail soak [Homo sapiens],6DIM_B Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT1982 from cocktail soak [Homo sapiens],6DJD_A Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT1982 (single soak) [Homo sapiens],6DJD_B Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT1982 (single soak) [Homo sapiens],6DJE_A Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound CDS010292 [Homo sapiens],6DJE_B Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound CDS010292 [Homo sapiens],6DJF_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ502 [Homo sapiens],6DJF_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ502 [Homo sapiens],6DJG_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ503 [Homo sapiens],6DJG_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ503 [Homo sapiens],6DJH_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ515 [Homo sapiens],6DJH_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ515 [Homo sapiens],6DJI_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ522 [Homo sapiens],6DJI_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ522 [Homo sapiens],6DJJ_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ532 [Homo sapiens],6DJJ_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ532 [Homo sapiens],6MYZ_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ520 [Homo sapiens],6MYZ_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ520 [Homo sapiens],6MZ0_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ530 [Homo sapiens],6MZ0_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ530 [Homo sapiens],6N0D_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ575 [Homo sapiens],6N0D_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ575 [Homo sapiens],6N0N_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ574 [Homo sapiens],6N0N_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ574 [Homo sapiens],6N0O_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ523 [Homo sapiens],6N0O_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ523 [Homo sapiens],6N0R_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ572 [Homo sapiens],6N0R_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ572 [Homo sapiens],6N17_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ577 [Homo sapiens],6N17_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ577 [Homo sapiens],6N19_A Crystal structure of Tdp1 catalytic domain in complex with compound XZ578 [Homo sapiens],6N19_B Crystal structure of Tdp1 catalytic domain in complex with compound XZ578 [Homo sapiens],6W4R_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],6W4R_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],6W7J_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],6W7J_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],6W7K_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],6W7K_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],6W7L_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],6W7L_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens] |
|
| 1.92e-56 | 703 | 1189 | 37 | 480 | human Tyrosyl DNA phosphodiesterase [Homo sapiens],1QZQ_B human Tyrosyl DNA phosphodiesterase [Homo sapiens] |
|
| 5.00e-56 | 703 | 1189 | 39 | 482 | Chain A, Tyrosyl-DNA Phosphodiesterase [Homo sapiens],1MU7_B Chain B, Tyrosyl-DNA Phosphodiesterase [Homo sapiens],1MU9_A Chain A, Tyrosyl-DNA Phosphodiesterase [Homo sapiens],1MU9_B Chain B, Tyrosyl-DNA Phosphodiesterase [Homo sapiens],1NOP_A Chain A, tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1NOP_B Chain B, tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RFF_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RFF_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RFI_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RFI_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RG1_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RG1_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RG2_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RG2_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RGT_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RGT_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RGU_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RGU_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RH0_A Chain A, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],1RH0_B Chain B, Tyrosyl-DNA phosphodiesterase 1 [Homo sapiens],5NW9_A Crystal structure of the complex of Tdp1 with duplex DNA [Homo sapiens],5NW9_B Crystal structure of the complex of Tdp1 with duplex DNA [Homo sapiens],5NWA_A Crystal structure of the complex of Tdp1 with duplex DNA [Homo sapiens],5NWA_B Crystal structure of the complex of Tdp1 with duplex DNA [Homo sapiens] |
|
| 2.18e-54 | 703 | 1189 | 18 | 461 | Chain A, TYROSYL-DNA PHOSPHODIESTERASE [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.09e-296 | 4 | 605 | 5 | 613 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=alg10 PE=3 SV=1 |
|
| 6.91e-280 | 1 | 605 | 1 | 607 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=alg10 PE=3 SV=1 |
|
| 2.31e-97 | 27 | 605 | 60 | 659 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=ALG10 PE=3 SV=1 |
|
| 3.58e-89 | 2 | 605 | 43 | 721 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ALG10 PE=3 SV=1 |
|
| 3.15e-75 | 27 | 605 | 61 | 770 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-10 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.019907 | 0.980058 | CS pos: 33-34. Pr: 0.9405 |
TMHMM Annotations download full data without filtering help
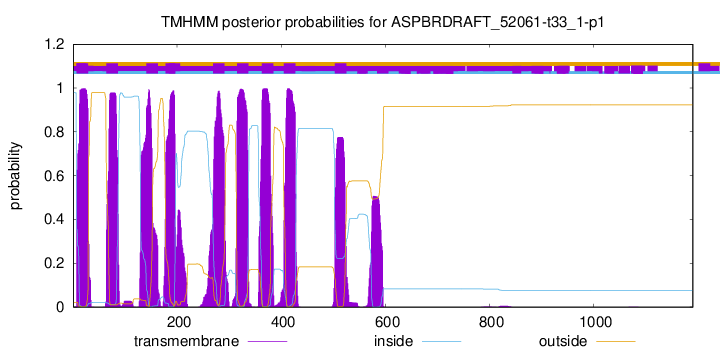
| Start | End |
|---|---|
| 7 | 29 |
| 64 | 86 |
| 141 | 163 |
| 178 | 196 |
| 269 | 291 |
| 315 | 337 |
| 363 | 385 |
| 405 | 427 |
| 503 | 525 |
