You are browsing environment: FUNGIDB
CAZyme Information: ASPBRDRAFT_45473-t33_1-p1
You are here: Home > Sequence: ASPBRDRAFT_45473-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus brasiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus brasiliensis | |||||||||||
| CAZyme ID | ASPBRDRAFT_45473-t33_1-p1 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 43 | 171 | 8.3e-21 | 0.5814977973568282 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 4.81e-17 | 42 | 267 | 46 | 304 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 226584 | COG4099 | 2.87e-07 | 45 | 226 | 176 | 363 | Predicted peptidase [General function prediction only]. |
| 223489 | DLH | 4.02e-06 | 118 | 194 | 96 | 173 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 223477 | YpfH | 0.002 | 113 | 205 | 78 | 172 | Predicted esterase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.04e-90 | 26 | 267 | 38 | 285 | |
| 2.12e-89 | 22 | 267 | 92 | 352 | |
| 7.17e-84 | 22 | 267 | 42 | 302 | |
| 1.48e-83 | 10 | 267 | 10 | 272 | |
| 3.61e-83 | 22 | 267 | 18 | 279 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.29e-200 | 1 | 270 | 1 | 270 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
|
| 3.25e-169 | 1 | 270 | 1 | 270 | Feruloyl esterase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=faeC PE=1 SV=1 |
|
| 3.69e-159 | 1 | 270 | 1 | 270 | Probable feruloyl esterase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=faeC PE=3 SV=1 |
|
| 1.23e-157 | 1 | 270 | 1 | 270 | Probable feruloyl esterase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=faeC PE=3 SV=2 |
|
| 2.19e-156 | 1 | 270 | 1 | 272 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
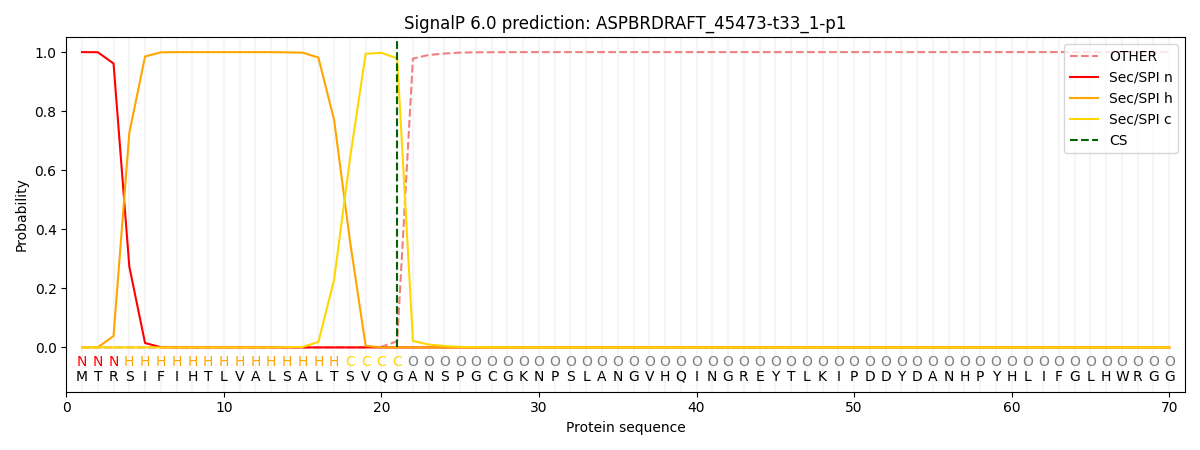
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000230 | 0.999729 | CS pos: 21-22. Pr: 0.9789 |
