You are browsing environment: FUNGIDB
CAZyme Information: ASPACDRAFT_35448-t33_1-p1
Basic Information
help
| Species |
Aspergillus aculeatus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus aculeatus
|
| CAZyme ID |
ASPACDRAFT_35448-t33_1-p1
|
| CAZy Family |
GH28 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 399 |
KV878987|CGC6 |
41404.83 |
4.6352 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AaculeatusATCC16872 |
11165 |
690307 |
322 |
10843
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| AA11 |
22 |
223 |
2.9e-73 |
0.9895287958115183 |
ASPACDRAFT_35448-t33_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 8.03e-108 |
2 |
385 |
1 |
387 |
| 4.70e-107 |
2 |
385 |
1 |
388 |
| 4.70e-107 |
2 |
385 |
1 |
388 |
| 9.43e-103 |
28 |
399 |
26 |
384 |
| 2.65e-98 |
28 |
394 |
26 |
388 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.95e-87 |
28 |
233 |
7 |
216 |
Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
ASPACDRAFT_35448-t33_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
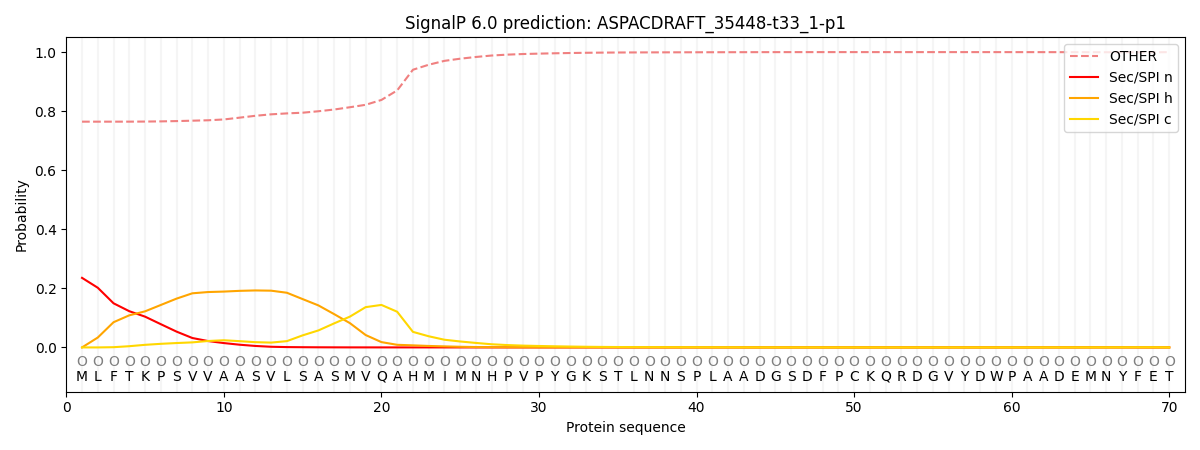
| Other |
SP_Sec_SPI |
CS Position |
| 0.775013 |
0.224982 |
|
There is no transmembrane helices in ASPACDRAFT_35448-t33_1-p1.

