You are browsing environment: FUNGIDB
CAZyme Information: ASPACDRAFT_1880973-t33_1-p1
You are here: Home > Sequence: ASPACDRAFT_1880973-t33_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus aculeatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus aculeatus | |||||||||||
| CAZyme ID | ASPACDRAFT_1880973-t33_1-p1 | |||||||||||
| CAZy Family | CE2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 57 | 475 | 1.3e-55 | 0.9694323144104804 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396238 | FAD_binding_4 | 2.02e-13 | 62 | 201 | 1 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 223354 | GlcD | 4.21e-13 | 62 | 270 | 32 | 240 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 178402 | PLN02805 | 9.32e-05 | 55 | 228 | 130 | 303 | D-lactate dehydrogenase [cytochrome] |
| 215242 | PLN02441 | 8.78e-04 | 10 | 223 | 15 | 232 | cytokinin dehydrogenase |
| 411067 | Arc1 | 0.004 | 336 | 410 | 1 | 62 | Arc1, AvrRxo1-required chaperone. This family contains AvrRxo1-ORF2 (also called AvrRxo1-required chaperone or Arc1) which appears to act as a molecular chaperone for AvrRxo1-ORF1 (also called AvrRxo1), a type III-secreted virulence factor in Xanthomonas oryzae (Xoc), a bacteria that causes leaf streak (BLS) disease in rice plants. AvrRxo1-ORF1 delivery in rice plant cells is recognized by disease resistance protein Rxo1, which triggers resistance to BLS disease. In the Xoc genome, the Arc1 gene is found adjacent to AvrRxo1; Arc1 functions to suppress the bacteriostatic activity of AvrRxo1-ORF1 in bacterial cells. Arc1 has a kinase-binding domain with a structure that is atypical of effector-binding chaperones, while AvrRxo1 has a T4 polynucleotide kinase (T4pnk) domain. AvrRxo1 and Arc1 comprise a toxin-antitoxin system similar to members of the zeta-epsilon family, with Arc1 acting as the antitoxin. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.58e-22 | 4 | 270 | 5 | 269 | |
| 1.14e-17 | 38 | 231 | 24 | 215 | |
| 1.14e-17 | 38 | 231 | 24 | 215 | |
| 3.65e-17 | 38 | 231 | 24 | 215 | |
| 1.13e-16 | 48 | 284 | 106 | 339 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.14e-17 | 35 | 226 | 19 | 205 | The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_B The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_C The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_D The crystal structure of EncM T139V mutant [Streptomyces maritimus] |
|
| 3.80e-17 | 35 | 226 | 19 | 205 | The crystal structure of EncM V135M mutant [Streptomyces maritimus],6FYF_B The crystal structure of EncM V135M mutant [Streptomyces maritimus],6FYF_C The crystal structure of EncM V135M mutant [Streptomyces maritimus],6FYF_D The crystal structure of EncM V135M mutant [Streptomyces maritimus] |
|
| 3.80e-17 | 35 | 226 | 19 | 205 | The crystal structure of EncM V135T mutant [Streptomyces maritimus],6FYG_B The crystal structure of EncM V135T mutant [Streptomyces maritimus],6FYG_C The crystal structure of EncM V135T mutant [Streptomyces maritimus],6FYG_D The crystal structure of EncM V135T mutant [Streptomyces maritimus] |
|
| 5.07e-17 | 35 | 226 | 19 | 205 | The crystal structure of EncM H138T mutant [Streptomyces maritimus],6FYE_B The crystal structure of EncM H138T mutant [Streptomyces maritimus] |
|
| 6.77e-17 | 35 | 226 | 19 | 205 | The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYB_B The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYB_C The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYB_D The crystal structure of EncM L144M mutant [Streptomyces maritimus],6FYC_A The crystal structure of EncM L144M mutant complex with dioxygen under 15 bars O2 pressure [Streptomyces maritimus],6FYC_B The crystal structure of EncM L144M mutant complex with dioxygen under 15 bars O2 pressure [Streptomyces maritimus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.06e-159 | 25 | 483 | 8 | 476 | FAD-dependent monooxygenase sdcF OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=sdcF PE=1 SV=1 |
|
| 7.04e-123 | 25 | 488 | 44 | 518 | FAD-dependent monooxygenase drtC OS=Aspergillus calidoustus OX=454130 GN=drtC PE=1 SV=1 |
|
| 1.41e-76 | 36 | 484 | 52 | 518 | FAD-dependent monooxygenase tpcD OS=Cochliobolus heterostrophus (strain C5 / ATCC 48332 / race O) OX=701091 GN=tpcD PE=1 SV=1 |
|
| 3.03e-73 | 17 | 483 | 36 | 524 | FAD-dependent monooxygenase CTB5 OS=Cercospora beticola OX=122368 GN=CTB5 PE=3 SV=1 |
|
| 1.41e-64 | 52 | 483 | 3 | 456 | FAD-dependent monooxygenase CTB5 OS=Cercospora nicotianae OX=29003 GN=CTB5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
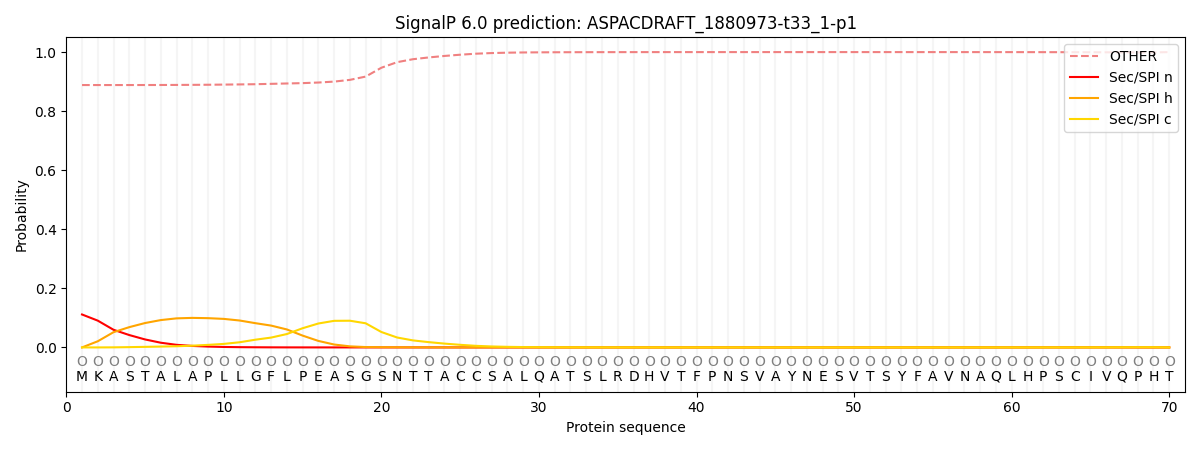
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.896204 | 0.103825 |
