You are browsing environment: FUNGIDB
CAZyme Information: ASPACDRAFT_1870924-t33_1-p1
Basic Information
help
| Species |
Aspergillus aculeatus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus aculeatus
|
| CAZyme ID |
ASPACDRAFT_1870924-t33_1-p1
|
| CAZy Family |
CBM21 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1024 |
KV878979|CGC6 |
110423.66 |
5.1943 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AaculeatusATCC16872 |
11165 |
690307 |
322 |
10843
|
|
| Gene Location |
No EC number prediction in ASPACDRAFT_1870924-t33_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
34 |
780 |
1.7e-75 |
0.7851941747572816 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 407266
|
Glyco_hydro_106 |
1.09e-17 |
302 |
744 |
434 |
869 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
21 |
1010 |
21 |
999 |
| 0.0 |
21 |
1010 |
21 |
999 |
| 0.0 |
12 |
1014 |
12 |
987 |
| 0.0 |
4 |
1014 |
3 |
996 |
| 0.0 |
8 |
1014 |
8 |
1018 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 3.96e-09 |
307 |
966 |
503 |
1096 |
Structure of Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 3.09e-07 |
283 |
627 |
275 |
601 |
Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
This protein is predicted as SP
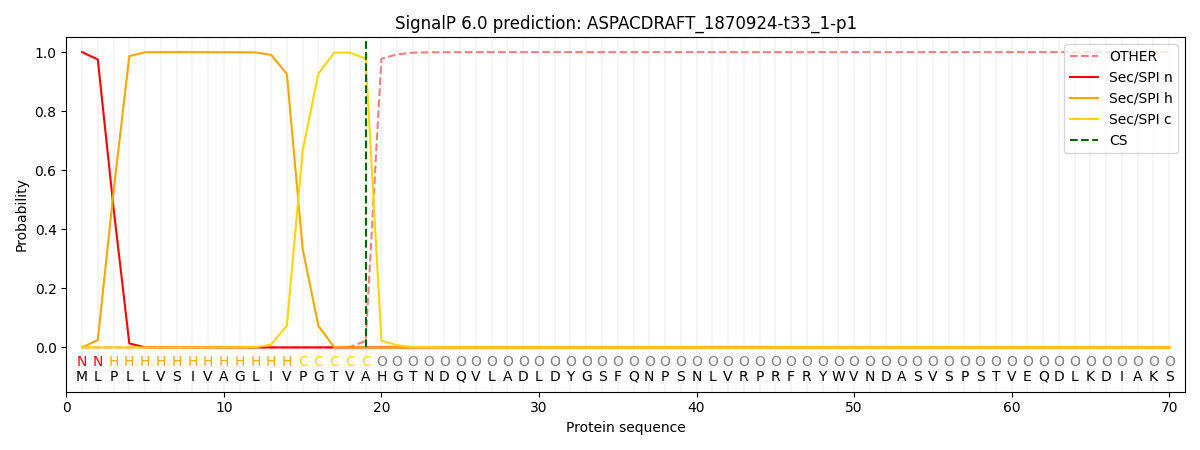
| Other |
SP_Sec_SPI |
CS Position |
| 0.000244 |
0.999719 |
CS pos: 19-20. Pr: 0.9779 |
There is no transmembrane helices in ASPACDRAFT_1870924-t33_1-p1.

