You are browsing environment: FUNGIDB
CAZyme Information: APA15443.1
You are here: Home > Sequence: APA15443.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sclerotinia sclerotiorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Sclerotiniaceae; Sclerotinia; Sclerotinia sclerotiorum | |||||||||||
| CAZyme ID | APA15443.1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.59:2 | - | - |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH71 | 180 | 569 | 2.7e-147 | 0.9946666666666667 |
| CBM24 | 610 | 685 | 1.9e-28 | 0.9868421052631579 |
| CBM24 | 704 | 780 | 4.1e-24 | 0.9605263157894737 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397634 | Glyco_hydro_71 | 0.0 | 180 | 568 | 1 | 372 | Glycosyl hydrolase family 71. Family of alpha-1,3-glucanases. |
| 211418 | GH71 | 6.76e-136 | 175 | 457 | 2 | 283 | Glycoside hydrolase family 71. This family of glycoside hydrolases 71 (following the CAZY nomenclature) function as alpha-1,3-glucanases (mutanases, EC 3.2.1.59). They appear to have an endo-hydrolytic mode of enzymatic activity and bacterial members are investigated as candidates for the development of dental caries treatments.The member from fission yeast, endo-alpha-1,3-glucanase Agn1p, plays a vital role in daughter cell separation, while Agn2p has been associated with endolysis of the ascus wall. |
| 211414 | GH99_GH71_like | 1.48e-44 | 180 | 451 | 1 | 283 | Glycoside hydrolase families 71, 99, and related domains. This superfamily of glycoside hydrolases contains families GH71 and GH99 (following the CAZY nomenclature), as well as other members with undefined function and specificity. |
| 211416 | GH99_GH71_like_3 | 4.70e-09 | 255 | 421 | 142 | 312 | Uncharacterized glycoside hydrolase family 99-like domain. This family of putative glycoside hydrolases resembles glycosyl hydrolase families 71 and 99 (following the CAZY nomenclature) and may share a similar catalytic site and mechanism. |
| 227709 | ROM1 | 0.001 | 32 | 178 | 135 | 269 | RhoGEF, Guanine nucleotide exchange factor for Rho/Rac/Cdc42-like GTPases [Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1272 | 1 | 1272 | |
| 2.39e-207 | 185 | 976 | 1 | 808 | |
| 2.24e-199 | 179 | 918 | 21 | 761 | |
| 5.45e-199 | 174 | 661 | 75 | 560 | |
| 5.45e-199 | 174 | 661 | 75 | 560 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.66e-62 | 169 | 594 | 27 | 432 | Mutanase Pc12g07500 OS=Penicillium rubens (strain ATCC 28089 / DSM 1075 / NRRL 1951 / Wisconsin 54-1255) OX=500485 GN=PCH_Pc12g07500 PE=1 SV=1 |
|
| 1.79e-61 | 178 | 590 | 22 | 420 | Glucan endo-1,3-alpha-glucosidase agn1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn1 PE=1 SV=2 |
|
| 7.57e-33 | 168 | 482 | 1 | 330 | Ascus wall endo-1,3-alpha-glucanase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
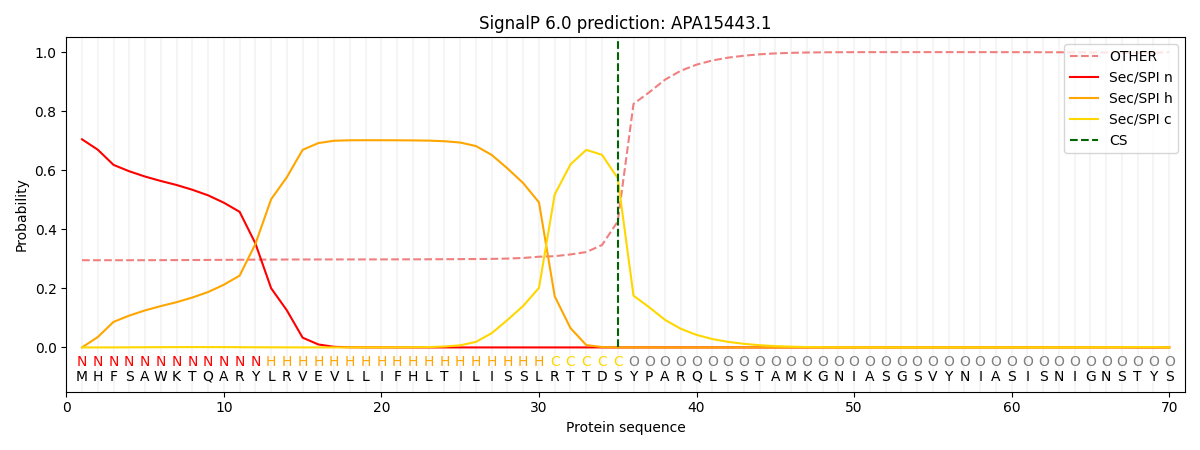
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.313402 | 0.686571 | CS pos: 35-36. Pr: 0.5731 |
