You are browsing environment: FUNGIDB
CAZyme Information: APA06419.1
You are here: Home > Sequence: APA06419.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sclerotinia sclerotiorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Sclerotiniaceae; Sclerotinia; Sclerotinia sclerotiorum | |||||||||||
| CAZyme ID | APA06419.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 171 | 392 | 6e-60 | 0.9776785714285714 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 371727 | Glyco_hydro_cc | 2.67e-69 | 161 | 392 | 2 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| 396406 | WSC | 1.11e-25 | 39 | 122 | 2 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 214616 | WSC | 1.63e-20 | 34 | 132 | 1 | 95 | present in yeast cell wall integrity and stress response component proteins. Domain present in WSC proteins, polycystin and fungal exoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.40e-305 | 1 | 394 | 1 | 394 | |
| 1.20e-211 | 5 | 394 | 83 | 467 | |
| 1.20e-211 | 5 | 394 | 83 | 467 | |
| 2.85e-210 | 1 | 394 | 1 | 390 | |
| 5.14e-122 | 149 | 394 | 22 | 269 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.72e-36 | 148 | 392 | 2 | 227 | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -2 and -1 subsites [Blastomyces gilchristii SLH14081] |
|
| 3.83e-36 | 148 | 392 | 33 | 258 | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) [Blastomyces gilchristii],6UAY_B Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) [Blastomyces gilchristii],6UAZ_A Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with glucose [Blastomyces gilchristii],6UAZ_B Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with glucose [Blastomyces gilchristii],6UB1_A Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -3 and -2 subsites [Blastomyces gilchristii],6UB1_B Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -3 and -2 subsites [Blastomyces gilchristii],6UB1_C Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -3 and -2 subsites [Blastomyces gilchristii],6UB1_D Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -3 and -2 subsites [Blastomyces gilchristii] |
|
| 2.62e-11 | 176 | 393 | 219 | 437 | Chain A, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans] |
|
| 9.34e-11 | 149 | 372 | 30 | 251 | Chain A, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.21e-24 | 5 | 130 | 35 | 158 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
|
| 3.63e-17 | 146 | 394 | 292 | 530 | Alkali-sensitive linkage protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=asl1 PE=1 SV=1 |
|
| 1.41e-09 | 36 | 131 | 18 | 109 | Putative fungistatic metabolite OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=CHGG_05463 PE=1 SV=2 |
|
| 8.92e-09 | 39 | 177 | 138 | 250 | WSC domain-containing protein 2 OS=Mus musculus OX=10090 GN=Wscd2 PE=2 SV=1 |
|
| 1.18e-08 | 39 | 177 | 139 | 251 | WSC domain-containing protein 2 OS=Danio rerio OX=7955 GN=wscd2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
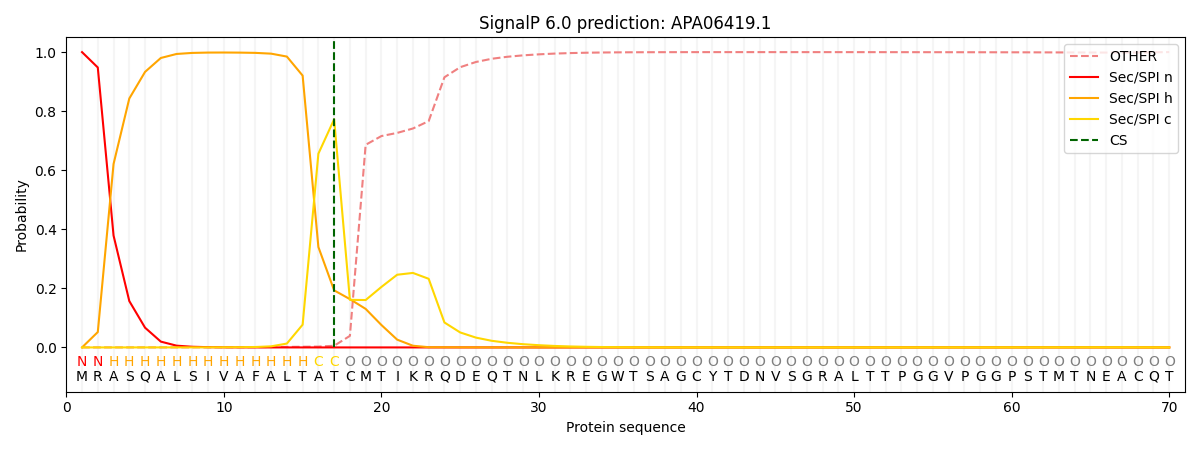
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000599 | 0.999388 | CS pos: 17-18. Pr: 0.7725 |
