You are browsing environment: FUNGIDB
CAZyme Information: AO090103000043-T-p1
You are here: Home > Sequence: AO090103000043-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus oryzae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus oryzae | |||||||||||
| CAZyme ID | AO090103000043-T-p1 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | Has domain(s) with predicted hydrolase activity, hydrolyzing O-glycosyl compounds, oxidoreductase activity and role in carbohydrate metabolic process, oxidation-reduction process | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1151378; End:1155376 Strand: + | |||||||||||
Full Sequence Download help
| MDFIAACPRR KCFYWKAYIA VPAALGSYHQ RRLLGCSIPN RPEILVVVWF WIMCIILNFG | 60 |
| FHDIFIPNVT MPTISRQAWK YVAQRTSMFA YACLPWVWLF AGRNNIFIWA TGWSFSTFNV | 120 |
| FHRHLSRLTA IFAFVHAISY TVLDTIYGPY YEEGLHVLWF KFGIIGATMM GLLCGLSIPY | 180 |
| IRTKCYELFL VGHILFAVIL LVALFQCVLL CSLPVYDFGR CETGLQDKTD ILPFSELNIT | 240 |
| PTSGQWLPYG AVIGSCASYA SEEADVVRLD VQADGSIRPL PGHYFHLYQL FSWRGWENHP | 300 |
| FTVGAYTAPN PTSPLTSSSN AQQGPHYMTG ALNEKNPFTS TPTEVFTQGS QSSTTDPALG | 360 |
| NLVFWIRPYN GWTKRLKDQC LRSSSSDDSA VNGYYQTSIP ISLEGPYGHT LPLHHYGTVV | 420 |
| MIVGGTGIAT AVPYIHDHVS RLSKAGSRVS DDTKTQTSNL TLVWACRGHV FMQQLCGREL | 480 |
| AGALEIEGFT GRFHCTKGCN SHCAADNKNG ELRIERGRPN IENIIHGAAT EAQIARHKMA | 540 |
| VLTCGSPHMS DEVRRSVHRI LKKGYEGIEY FEEAYSWLSW PSHSFVAKLH LCYGLKITIP | 600 |
| KLTLSFGWYA PLFRGIDVTA FRDPYVFQNK DLDDTVGSSS GTWYAAISGG VHDVGPGVFL | 660 |
| YRNQGSGYED WEYLGKWWSE PVNSTCGNGD WAKAWGYNFE VGNVFSLDKE GYNVNGETFI | 720 |
| TLGVEGSYGP ITESVTSMHG MLWASGNILK PDGGNVTFEP TMAGVLDWGI SSYAAAGKVL | 780 |
| PATSQASEKS GAPDRFISYV WLTGDVFGGV AGYPSEQQGW QNTLLLPRKL HKQTISNVVD | 840 |
| NDLASETGSW CVKDSENSCL KLETMGIKIA RETYKAMTNK TSFTEPKRTF CEDGAVPFKQ | 900 |
| SPTTKFFVLN AQLSFPKSAR DPGVQAGFKI LSSELESTTI YYQFSNESIV IGRTQPSRPP | 960 |
| LKLNTTTESG RLRLFDINKN CNKSDDDDKQ EERKKKDAYR EHGRNRHNVK LAAEDESHIE | 1020 |
| TLDLTIVVDN AVLEVYANSR FALSTWARTW YANSTEISFF HNGEGEVTFS DISVSDGLYD | 1080 |
| AYPDRAR | 1087 |
Enzyme Prediction help
| EC | 3.2.1.26:10 | 2.4.1.100:6 | 2.4.1.-:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH32 | 617 | 833 | 7.6e-17 | 0.5392491467576792 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350133 | GH32_XdINV-like | 1.50e-87 | 535 | 834 | 81 | 337 | glycoside hydrolase family 32 protein such as Xanthophyllomyces dendrorhous beta-fructofuranosidase (Inv;Xd-INV;XdINV). This subfamily of glycosyl hydrolase family GH32 includes fructan:fructan 1-fructosyltransferase (FT, EC 2.4.1.100) and beta-fructofuranosidase (invertase or Inv, EC 3.2.1.26), among others. These enzymes cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. Xanthophyllomyces dendrorhous beta-fructofuranosidase (XdINV) also catalyzes the synthesis of fructooligosaccharides (FOS, a beneficial prebiotic), producing neo-FOS, making it an interesting biotechnology target. Structural studies show plasticity of its active site, having a flexible loop that is essential in binding sucrose and beta(2-1)-linked oligosaccharide, making it a valuable biocatalyst to produce novel bioconjugates. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 99783 | NOX_Duox_like_FAD_NADP | 5.34e-35 | 265 | 575 | 11 | 210 | NADPH oxidase (NOX) catalyzes the generation of reactive oxygen species (ROS) such as superoxide and hydrogen peroxide. ROS were originally identified as bactericidal agents in phagocytes, but are now also implicated in cell signaling and metabolism. NOX has a 6-alpha helix heme-binding transmembrane domain fused to a flavoprotein with the nucleotide binding domain located in the cytoplasm. Duox enzymes link a peroxidase domain to the NOX domain via a single transmembrane and EF-hand Ca2+ binding sites. The flavoprotein module has a ferredoxin like FAD/NADPH binding domain. In classical phagocytic NOX2, electron transfer occurs from NADPH to FAD to the heme of cytb to oxygen leading to superoxide formation. |
| 396386 | Ferric_reduct | 2.51e-33 | 90 | 203 | 2 | 117 | Ferric reductase like transmembrane component. This family includes a common region in the transmembrane proteins mammalian cytochrome B-245 heavy chain (gp91-phox), ferric reductase transmembrane component in yeast and respiratory burst oxidase from mouse-ear cress. This may be a family of flavocytochromes capable of moving electrons across the plasma membrane. The Frp1 protein from S. pombe is a ferric reductase component and is required for cell surface ferric reductase activity, mutants in frp1 are deficient in ferric iron uptake. Cytochrome B-245 heavy chain is a FAD-dependent dehydrogenase it is also has electron transferase activity which reduces molecular oxygen to superoxide anion, a precursor in the production of microbicidal oxidants. Mutations in the sequence of cytochrome B-245 heavy chain (gp91-phox) lead to the X-linked chronic granulomatous disease. The bacteriocidal ability of phagocytic cells is reduced and is characterized by the absence of a functional plasma membrane associated NADPH oxidase. The chronic granulomatous disease gene codes for the beta chain of cytochrome B-245 and cytochrome B-245 is missing from patients with the disease. |
| 400517 | Glyco_hydro_32C | 8.10e-23 | 872 | 1076 | 1 | 162 | Glycosyl hydrolases family 32 C terminal. This domain corresponds to the C terminal domain of glycosyl hydrolase family 32. It forms a beta sandwich module. |
| 350110 | GH32_FFase | 2.05e-18 | 615 | 830 | 118 | 279 | Glycosyl hydrolase family 32, beta-fructosidases. Glycosyl hydrolase family GH32 cleaves sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). This family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAE65527.1|GH32 | 0.0 | 1 | 1087 | 1 | 1087 |
| QRD92928.1|GH32 | 0.0 | 1 | 1087 | 13 | 1088 |
| QMW36104.1|GH32 | 0.0 | 10 | 1087 | 124 | 1114 |
| QMW48164.1|GH32 | 0.0 | 10 | 1087 | 124 | 1114 |
| UDD65327.1|GH32 | 0.0 | 261 | 1087 | 5 | 753 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XH8_A | 3.61e-171 | 617 | 1085 | 165 | 603 | Aspergillus kawachii beta-fructofuranosidase complexed with glycerol [Aspergillus luchuensis IFO 4308],5XH9_A Aspergillus kawachii beta-fructofuranosidase [Aspergillus luchuensis IFO 4308],5XHA_A Aspergillus kawachii beta-fructofuranosidase complexed with fructose [Aspergillus luchuensis IFO 4308] |
| 3LF7_A | 1.93e-149 | 615 | 1085 | 164 | 634 | Crystal structure of fructosyltransferase (wild-type) from A. japonicus [Aspergillus japonicus],3LFI_A Crystal structure of fructosyltransferase (wild-type) from A. japonicus in complex with glucose [Aspergillus japonicus],3LFI_B Crystal structure of fructosyltransferase (wild-type) from A. japonicus in complex with glucose [Aspergillus japonicus] |
| 3LDK_A | 2.92e-148 | 615 | 1085 | 164 | 634 | Crystal Structure of A. japonicus CB05 [Aspergillus japonicus],3LDR_A Crystal structure of fructosyltransferase (D191A) from A. japonicus in complex with 1-Kestose [Aspergillus japonicus],3LEM_A Crystal structure of fructosyltransferase (D191A) from A. japonicus in complex with Nystose [Aspergillus japonicus],3LIG_A Crystal structure of fructosyltransferase (D191A) from A. japonicus [Aspergillus japonicus],3LIH_A Crystal structure of fructosyltransferase (D191A) from A. japonicus in complex with raffinose [Aspergillus japonicus] |
| 5FK7_A | 2.37e-36 | 617 | 1085 | 213 | 639 | Chain A, Beta-fructofuranosidase [Phaffia rhodozyma],5FK7_B Chain B, Beta-fructofuranosidase [Phaffia rhodozyma],5FK8_A Chain A, Beta-fructofuranosidase [Phaffia rhodozyma],5FK8_B Chain B, Beta-fructofuranosidase [Phaffia rhodozyma],5FKB_A Chain A, BETA-FRUCTOFURANOSIDASE [Phaffia rhodozyma],5FKB_B Chain B, BETA-FRUCTOFURANOSIDASE [Phaffia rhodozyma],5FKC_A Chain A, BETA-FRUCTOFURANOSIDASE [Phaffia rhodozyma],5FKC_B Chain B, BETA-FRUCTOFURANOSIDASE [Phaffia rhodozyma],5FMC_A Chain A, Beta-fructofuranosidase [Phaffia rhodozyma],5FMC_B Chain B, Beta-fructofuranosidase [Phaffia rhodozyma] |
| 5ANN_A | 2.41e-36 | 617 | 1085 | 215 | 641 | Chain A, Beta-fructofuranosidase [Phaffia rhodozyma],5ANN_B Chain B, Beta-fructofuranosidase [Phaffia rhodozyma] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q4WR75|FREB_ASPFU | 1.40e-94 | 13 | 577 | 200 | 743 | Ferric/cupric reductase transmembrane component B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=freB PE=2 SV=2 |
| sp|P36033|FRE2_YEAST | 4.66e-26 | 14 | 577 | 204 | 711 | Ferric/cupric reductase transmembrane component 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FRE2 PE=1 SV=1 |
| sp|Q08905|FRE3_YEAST | 1.42e-25 | 14 | 577 | 204 | 711 | Ferric reductase transmembrane component 3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FRE3 PE=1 SV=1 |
| sp|P53746|FRE4_YEAST | 1.67e-20 | 14 | 577 | 197 | 719 | Ferric reductase transmembrane component 4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FRE4 PE=2 SV=1 |
| sp|P32791|FRE1_YEAST | 3.90e-18 | 16 | 577 | 183 | 686 | Ferric/cupric reductase transmembrane component 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FRE1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999963 | 0.000024 |
TMHMM Annotations download full data without filtering help
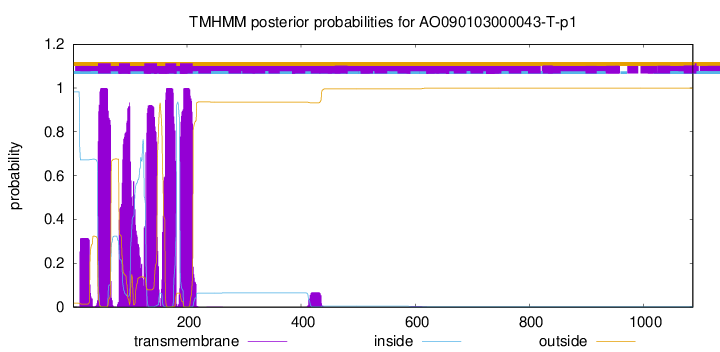
| Start | End |
|---|---|
| 44 | 66 |
| 81 | 100 |
| 125 | 147 |
| 162 | 181 |
| 188 | 210 |
