You are browsing environment: FUNGIDB
CAZyme Information: AO090038000131-T-p1
You are here: Home > Sequence: AO090038000131-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus oryzae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus oryzae | |||||||||||
| CAZyme ID | AO090038000131-T-p1 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | Ortholog of A. nidulans FGSC A4 : AN2537, A. fumigatus Af293 : Afu3g14890, Neosartorya fischeri NRRL 181 : NFIA_062360 and Aspergillus versicolor : Aspve1_0524633 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 45 | 407 | 9e-157 | 0.9942363112391931 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 271210 | PL-6 | 3.61e-04 | 70 | 122 | 8 | 59 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| 404168 | Beta_helix | 4.84e-04 | 160 | 326 | 21 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| 405302 | Chondroitinas_B | 0.001 | 69 | 122 | 7 | 59 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.62e-310 | 1 | 419 | 1 | 419 | |
| 4.62e-310 | 1 | 419 | 1 | 419 | |
| 2.97e-288 | 28 | 419 | 1 | 392 | |
| 2.97e-288 | 28 | 419 | 1 | 392 | |
| 2.97e-288 | 28 | 419 | 1 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.84e-49 | 49 | 417 | 18 | 380 | Chain A, Pectate lyase [Dickeya chrysanthemi] |
|
| 1.81e-21 | 49 | 332 | 6 | 321 | Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.53e-48 | 49 | 417 | 43 | 405 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
|
| 2.53e-48 | 49 | 417 | 43 | 405 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
|
| 9.73e-28 | 42 | 337 | 385 | 646 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
|
| 2.21e-10 | 48 | 338 | 36 | 333 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
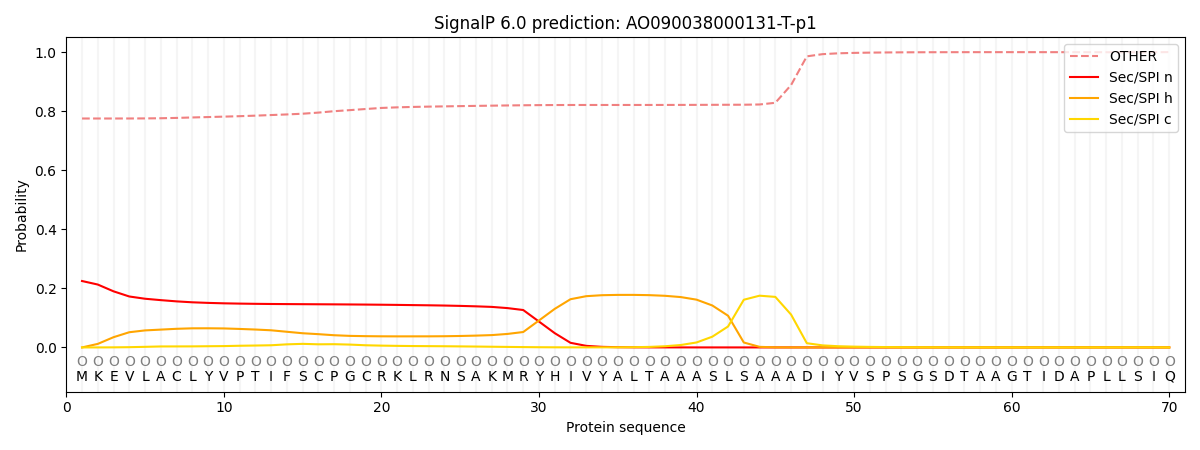
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.780534 | 0.219474 |
