You are browsing environment: FUNGIDB
AO090012000032-T-p1
Basic Information
help
Species
Aspergillus oryzae
Lineage
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus oryzae
CAZyme ID
AO090012000032-T-p1
CAZy Family
GH18|CBM19
CAZyme Description
Has domain(s) with predicted catalytic activity and role in carbohydrate metabolic process
CAZyme Property
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_AoryzaeRIB40
12364
510516
274
12090
Gene Location
No EC number prediction in AO090012000032-T-p1.
Family
Start
End
Evalue
family coverage
GH128
70
248
1.8e-25
0.7678571428571429
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
371727
Glyco_hydro_cc
9.55e-58
42
271
4
235
Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
6.07e-216
1
277
1
277
3.51e-215
1
277
1
277
8.89e-205
1
273
1
273
1.30e-112
1
277
1
272
3.66e-14
51
259
305
531
AO090012000032-T-p1 has no PDB hit.
AO090012000032-T-p1 has no Swissprot hit.
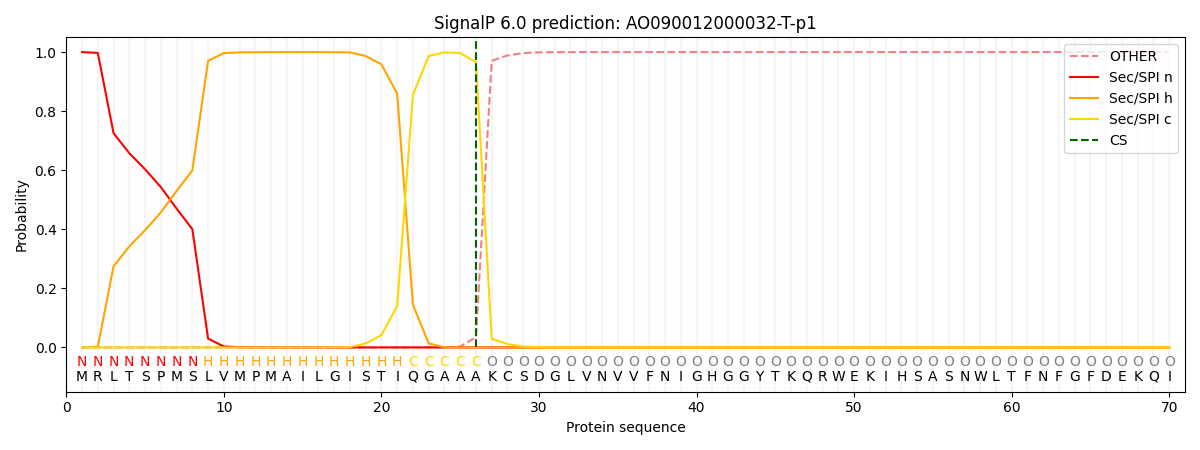
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000359
0.999624
CS pos: 26-27. Pr: 0.9657
There is no transmembrane helices in AO090012000032-T-p1.