You are browsing environment: FUNGIDB
CAZyme Information: AO090011000715-T-p1
You are here: Home > Sequence: AO090011000715-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus oryzae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus oryzae | |||||||||||
| CAZyme ID | AO090011000715-T-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | Ortholog(s) have cellulase activity, cellulose binding activity and role in xyloglucan catabolic process | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:38 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 33 | 309 | 4.4e-117 | 0.9964539007092199 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 1.19e-74 | 36 | 302 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| 225344 | BglC | 2.10e-24 | 31 | 331 | 45 | 391 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 197593 | fCBD | 3.28e-15 | 457 | 490 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 7.47e-14 | 458 | 486 | 1 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.53e-295 | 1 | 490 | 1 | 490 | |
| 4.18e-294 | 1 | 490 | 1 | 490 | |
| 4.18e-294 | 1 | 490 | 1 | 490 | |
| 4.18e-294 | 1 | 490 | 1 | 490 | |
| 2.41e-293 | 1 | 490 | 1 | 490 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.28e-156 | 31 | 330 | 3 | 303 | Structure of Thermoascus aurantiacus family 5 endoglucanase [Thermoascus aurantiacus],1GZJ_B Structure of Thermoascus aurantiacus family 5 endoglucanase [Thermoascus aurantiacus] |
|
| 3.81e-155 | 21 | 331 | 3 | 314 | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM [Talaromyces verruculosus],6T9F_B CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM [Talaromyces verruculosus],6T9F_C CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM [Talaromyces verruculosus],6T9F_D CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM [Talaromyces verruculosus] |
|
| 4.98e-155 | 31 | 330 | 2 | 301 | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger] |
|
| 2.24e-154 | 31 | 330 | 4 | 304 | Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus [Thermoascus aurantiacus],1H1N_B Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus [Thermoascus aurantiacus] |
|
| 4.35e-154 | 31 | 330 | 4 | 303 | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger],5I79_B Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.02e-158 | 5 | 330 | 6 | 331 | Probable endo-beta-1,4-glucanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=eglB PE=3 SV=1 |
|
| 5.06e-157 | 1 | 330 | 1 | 331 | Probable endo-beta-1,4-glucanase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=eglB PE=3 SV=1 |
|
| 5.06e-157 | 1 | 330 | 1 | 331 | probable endo-beta-1,4-glucanase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=eglB PE=3 SV=1 |
|
| 8.26e-155 | 1 | 330 | 1 | 327 | Probable endo-beta-1,4-glucanase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglB PE=3 SV=1 |
|
| 1.45e-153 | 5 | 330 | 6 | 330 | Endo-beta-1,4-glucanase B OS=Aspergillus niger OX=5061 GN=eglB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
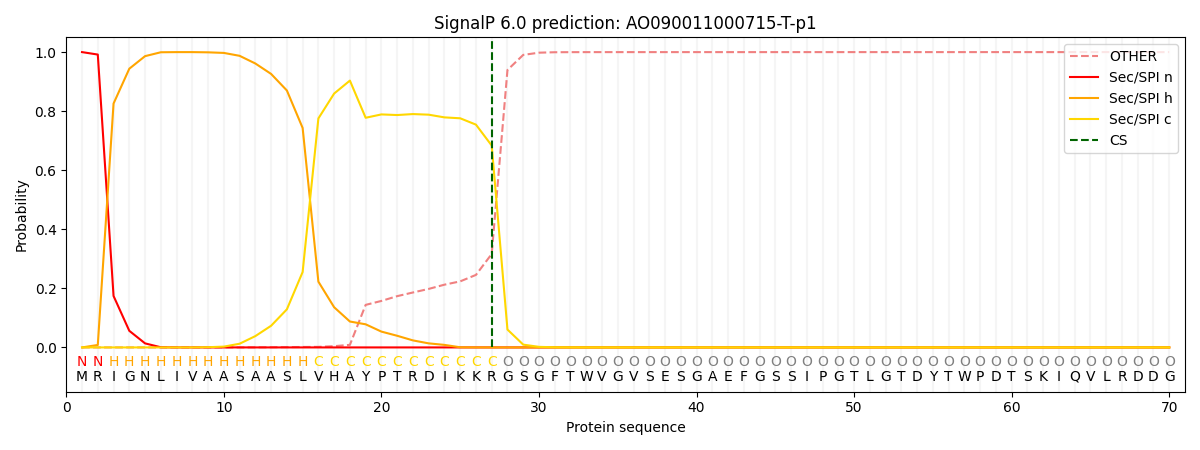
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000320 | 0.999660 | CS pos: 27-28. Pr: 0.6835 |
