You are browsing environment: FUNGIDB
AO090010000145-T-p1
Basic Information
help
Species
Aspergillus oryzae
Lineage
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus oryzae
CAZyme ID
AO090010000145-T-p1
CAZy Family
PL20
CAZyme Description
Ortholog of A. nidulans FGSC A4 : AN5663, A. fumigatus Af293 : Afu4g13710, A. niger CBS 513.88 : An01g09510, A. oryzae RIB40 : AO090009000200 and Aspergillus wentii : Aspwe1_0106727
CAZyme Property
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_AoryzaeRIB40
12364
510516
274
12090
Gene Location
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
215605
PLN03153
1.65e-14
233
359
212
341
hypothetical protein; Provisional
more
398362
DUF604
2.85e-11
277
457
4
204
Protein of unknown function, DUF604. This family includes a conserved region found in several uncharacterized plant proteins.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
0.0
1
483
1
483
0.0
1
483
1
483
0.0
1
483
1
483
0.0
1
483
1
483
2.65e-277
1
482
1
482
AO090010000145-T-p1 has no PDB hit.
AO090010000145-T-p1 has no Swissprot hit.
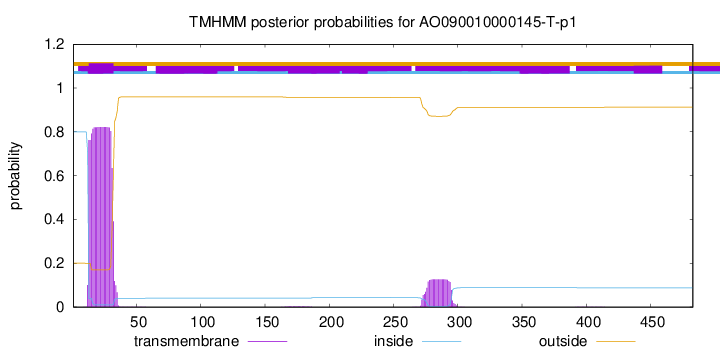
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
0.998465
0.001546