You are browsing environment: FUNGIDB
CAZyme Information: AO090003001140-T-p1
You are here: Home > Sequence: AO090003001140-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
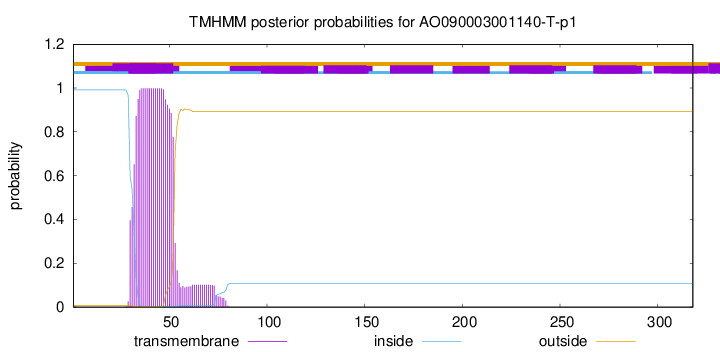
TMHMM annotations
Basic Information help
| Species | Aspergillus oryzae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus oryzae | |||||||||||
| CAZyme ID | AO090003001140-T-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | Ortholog(s) have alpha-1,6-mannosyltransferase activity, role in N-glycan processing and alpha-1,6-mannosyltransferase complex, endoplasmic reticulum localization | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 66 | 297 | 3.8e-78 | 0.9715447154471545 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 368535 | Glyco_transf_34 | 4.29e-72 | 63 | 292 | 1 | 232 | galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34. |
| 215616 | PLN03181 | 5.95e-04 | 88 | 148 | 155 | 211 | glycosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.24e-231 | 1 | 318 | 1 | 318 | |
| 1.24e-231 | 1 | 318 | 1 | 318 | |
| 1.24e-231 | 1 | 318 | 1 | 318 | |
| 1.24e-231 | 1 | 318 | 1 | 318 | |
| 1.24e-231 | 1 | 318 | 1 | 318 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.87e-40 | 82 | 318 | 123 | 357 | Uncharacterized alpha-1,2-galactosyltransferase C1289.13c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1289.13c PE=3 SV=1 |
|
| 3.62e-40 | 21 | 309 | 5 | 308 | Uncharacterized alpha-1,2-galactosyltransferase C8D2.17 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=pi048 PE=3 SV=2 |
|
| 8.17e-36 | 82 | 318 | 123 | 357 | Alpha-1,2-galactosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gma12 PE=1 SV=1 |
|
| 1.77e-31 | 67 | 309 | 160 | 417 | Probable alpha-1,6-mannosyltransferase MNN11 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN11 PE=1 SV=2 |
|
| 7.82e-29 | 47 | 307 | 67 | 327 | Alpha-1,2-galactosyltransferase gmh3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gmh3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999985 | 0.000016 |