You are browsing environment: FUNGIDB
CAZyme Information: AN4504-T-p1
Basic Information
help
| Species |
Aspergillus nidulans
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus nidulans
|
| CAZyme ID |
AN4504-T-p1
|
| CAZy Family |
GH28 |
| CAZyme Description |
Putative endo-mannanase GH76 family protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AnidulansFGSCA4 |
10988 |
227321 |
276 |
10712
|
|
| Gene Location |
No EC number prediction in AN4504-T-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH76 |
189 |
543 |
1.9e-56 |
0.7206703910614525 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 397638
|
Glyco_hydro_76 |
3.51e-15 |
269 |
524 |
120 |
313 |
Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 227170
|
COG4833 |
0.004 |
341 |
476 |
167 |
276 |
Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
631 |
1 |
631 |
| 1.93e-300 |
1 |
631 |
1 |
579 |
| 5.43e-217 |
56 |
629 |
71 |
619 |
| 5.43e-217 |
56 |
629 |
71 |
619 |
| 9.53e-217 |
56 |
629 |
57 |
605 |
AN4504-T-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 7.19e-08 |
278 |
493 |
156 |
337 |
Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=DCW1 PE=3 SV=1 |
This protein is predicted as SP
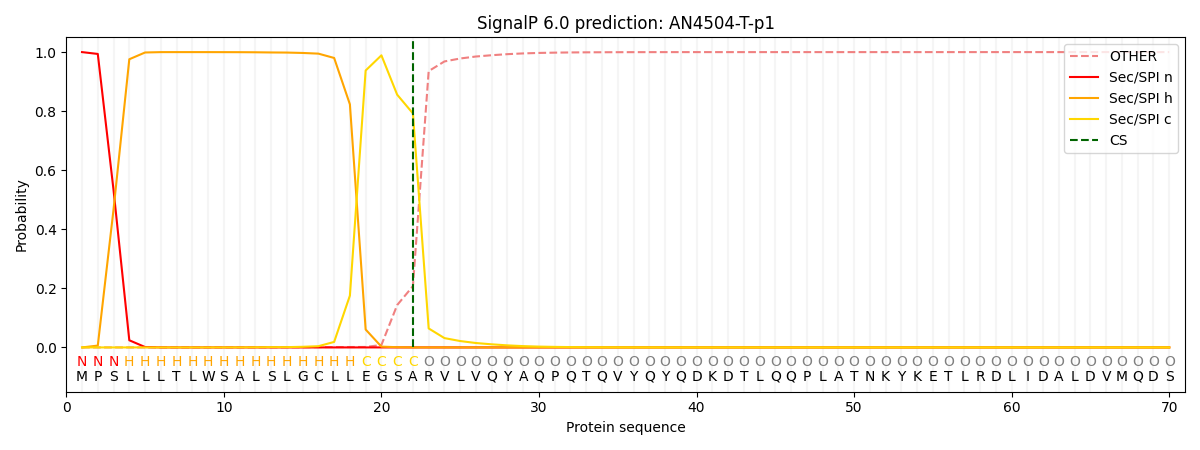
| Other |
SP_Sec_SPI |
CS Position |
| 0.000386 |
0.999597 |
CS pos: 22-23. Pr: 0.7923 |
There is no transmembrane helices in AN4504-T-p1.

