You are browsing environment: FUNGIDB
AN10041-T-p1
Basic Information
help
Species
Aspergillus nidulans
Lineage
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus nidulans
CAZyme ID
AN10041-T-p1
CAZy Family
AA6
CAZyme Description
Ortholog of A. niger CBS 513.88 : An07g01780, Neosartorya fischeri NRRL 181 : NFIA_113800, Aspergillus wentii : Aspwe1_0281884 and Aspergillus versicolor : Aspve1_0082092
CAZyme Property
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_AnidulansFGSCA4
10988
227321
276
10712
Gene Location
No EC number prediction in AN10041-T-p1.
Family
Start
End
Evalue
family coverage
GT34
67
191
7.9e-24
0.6016260162601627
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
368535
Glyco_transf_34
8.03e-10
63
188
21
170
galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34.
more
215617
PLN03182
1.66e-08
84
179
169
262
xyloglucan 6-xylosyltransferase; Provisional
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.04e-214
1
316
1
353
8.14e-109
5
311
7
351
1.43e-84
42
318
18
330
7.60e-81
51
264
51
308
1.55e-79
41
313
34
341
AN10041-T-p1 has no PDB hit.
AN10041-T-p1 has no Swissprot hit.
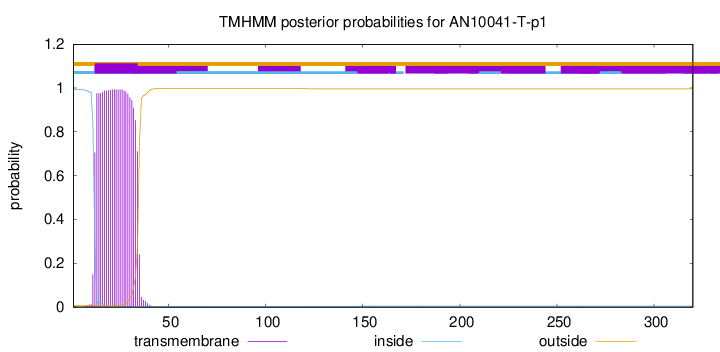
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.441208
0.558780
CS pos: 34-35. Pr: 0.2785