You are browsing environment: FUNGIDB
CAZyme Information: AN0550-T-p1
You are here: Home > Sequence: AN0550-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus nidulans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus nidulans | |||||||||||
| CAZyme ID | AN0550-T-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | Putative glucan 1,3-beta-glucosidase with a predicted role in glucan metabolism | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.58:16 | - | - |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 216 | 967 | 1.2e-281 | 0.9891891891891892 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403800 | Pectate_lyase_3 | 5.12e-97 | 244 | 470 | 1 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 403800 | Pectate_lyase_3 | 8.33e-10 | 605 | 721 | 1 | 125 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 227721 | Pgu1 | 1.23e-04 | 246 | 408 | 84 | 253 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1389 | 1 | 1380 | |
| 0.0 | 12 | 1381 | 12 | 1370 | |
| 0.0 | 56 | 1379 | 37 | 1355 | |
| 0.0 | 51 | 1383 | 47 | 1368 | |
| 0.0 | 59 | 1383 | 45 | 1355 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.31e-203 | 219 | 967 | 2 | 752 | Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
|
| 7.40e-193 | 223 | 967 | 26 | 755 | Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQN_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_A Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.28e-206 | 221 | 964 | 41 | 870 | Probable glucan endo-1,3-beta-glucosidase ARB_02077 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02077 PE=1 SV=1 |
|
| 2.28e-160 | 223 | 958 | 48 | 775 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
|
| 3.87e-56 | 211 | 937 | 26 | 726 | Glucan endo-1,3-beta-glucosidase BGN13.1 OS=Trichoderma harzianum OX=5544 GN=bgn13.1 PE=1 SV=1 |
|
| 1.47e-07 | 998 | 1375 | 39 | 421 | LysM domain-containing protein ARB_01155/01156 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_01155/01156 PE=3 SV=2 |
|
| 5.37e-07 | 968 | 1301 | 1 | 383 | LysM domain-containing protein ARB_03442 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_03442 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
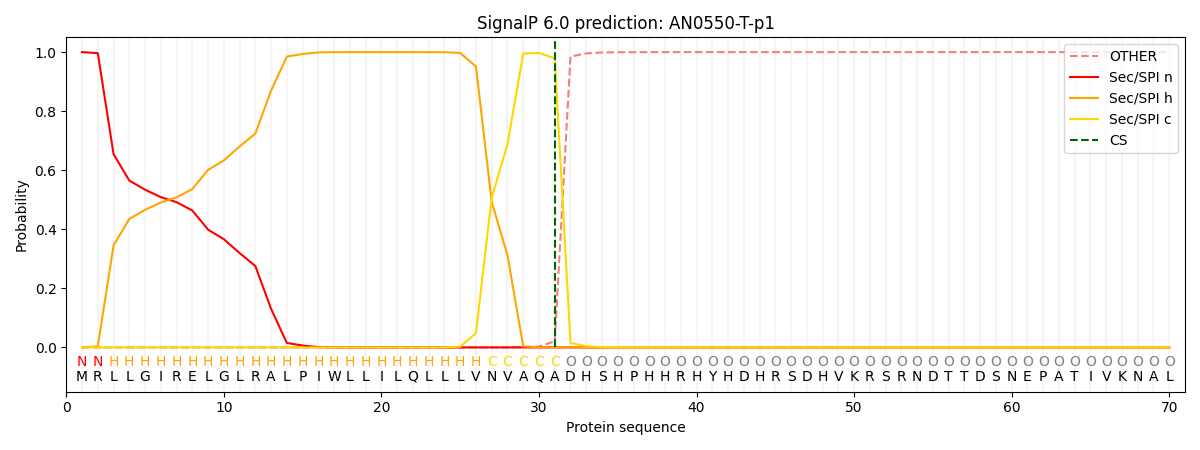
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000243 | 0.999714 | CS pos: 31-32. Pr: 0.9790 |
