You are browsing environment: FUNGIDB
CAZyme Information: AMAG_15319-t26_1-p1
You are here: Home > Sequence: AMAG_15319-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
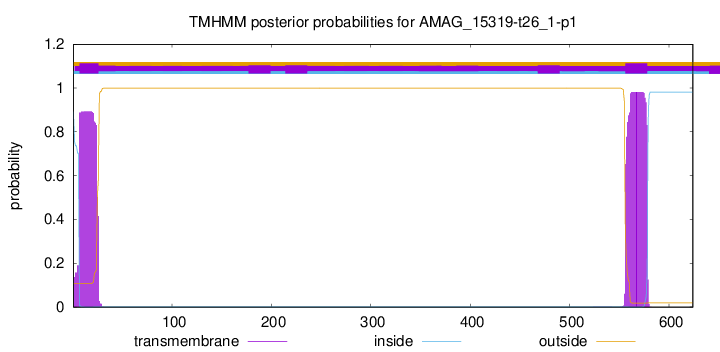
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_15319-t26_1-p1 | |||||||||||
| CAZy Family | GT24 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.214:5 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT10 | 50 | 369 | 1e-56 | 0.8847262247838616 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395683 | Glyco_transf_10 | 7.05e-31 | 197 | 382 | 4 | 169 | Glycosyltransferase family 10 (fucosyltransferase) C-term. This is the C-terminal domain of a family of fucosyltransferases. This enzyme transfers fucose from GDP-Fucose to GlcNAc in an alpha1,3 linkage. This family is known as glycosyltransferase family 10. The C-terminal domain is the likely binding-region for ADP (manuscript in publication). |
| 407214 | Glyco_tran_10_N | 1.93e-14 | 62 | 167 | 6 | 107 | Fucosyltransferase, N-terminal. This is the N-terminal domain of a family of fucosyltransferases. This enzyme transfers fucose from GDP-Fucose to GlcNAc in an alpha1,3 linkage. This family is known as glycosyltransferase family 10. The N-terminal domain is the likely binding-region for the fucose-like substrate (manuscript in publication). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.78e-27 | 152 | 363 | 21 | 228 | |
| 5.29e-24 | 62 | 335 | 93 | 374 | |
| 6.03e-24 | 62 | 331 | 93 | 370 | |
| 1.47e-23 | 84 | 331 | 81 | 338 | |
| 5.61e-23 | 62 | 331 | 80 | 357 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.97e-24 | 62 | 331 | 80 | 357 | Alpha-(1,3)-fucosyltransferase 11 OS=Oryzias latipes OX=8090 GN=fut11 PE=2 SV=1 |
|
| 3.28e-23 | 62 | 335 | 84 | 365 | Alpha-(1,3)-fucosyltransferase 11 OS=Takifugu rubripes OX=31033 GN=fut11 PE=2 SV=1 |
|
| 8.04e-23 | 59 | 378 | 86 | 421 | Alpha-(1,3)-fucosyltransferase 11 OS=Gallus gallus OX=9031 GN=FUT11 PE=2 SV=1 |
|
| 1.27e-21 | 27 | 383 | 84 | 456 | Alpha-(1,3)-fucosyltransferase 11 OS=Xenopus tropicalis OX=8364 GN=fut11 PE=2 SV=1 |
|
| 2.22e-21 | 84 | 331 | 82 | 339 | Alpha-(1,3)-fucosyltransferase 11 OS=Danio rerio OX=7955 GN=fut11 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
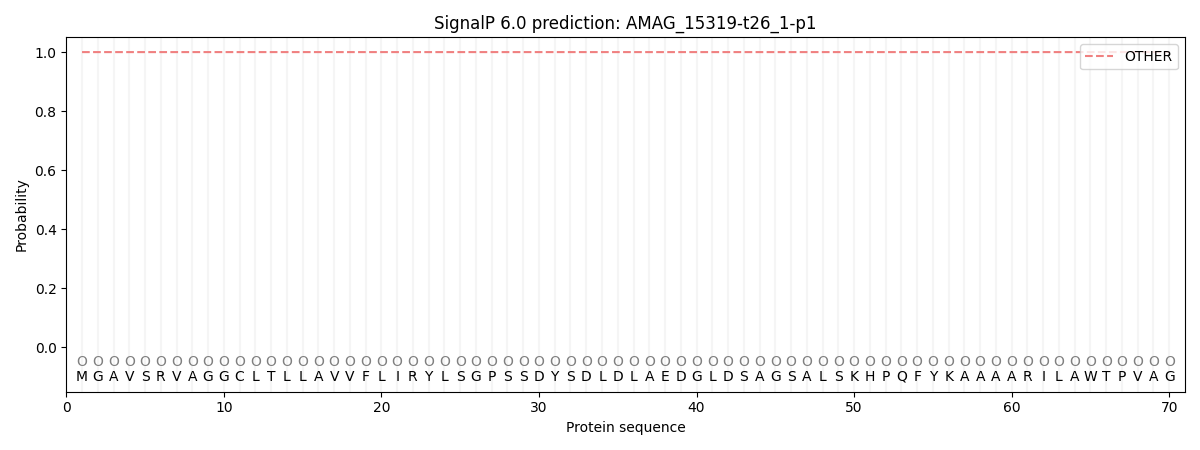
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000015 | 0.000007 |