You are browsing environment: FUNGIDB
CAZyme Information: AMAG_13480-t26_1-p1
You are here: Home > Sequence: AMAG_13480-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
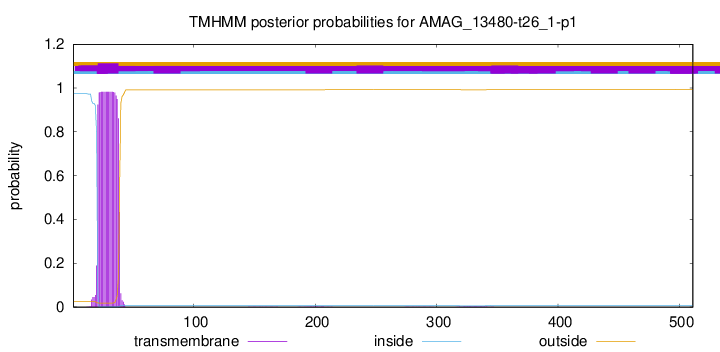
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_13480-t26_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.149:9 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT31 | 175 | 370 | 4.6e-37 | 0.9739583333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 6.31e-20 | 175 | 373 | 3 | 196 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 215596 | PLN03133 | 1.83e-09 | 141 | 419 | 373 | 626 | beta-1,3-galactosyltransferase; Provisional |
| 178735 | PLN03193 | 4.46e-04 | 159 | 370 | 140 | 348 | beta-1,3-galactosyltransferase; Provisional |
| 259799 | SPFH_prohibitin | 0.006 | 432 | 462 | 159 | 190 | Prohibitin family; SPFH (stomatin, prohibitin, flotillin, and HflK/C) superfamily. This model characterizes proteins similar to prohibitin (a lipid raft-associated integral membrane protein). Individual proteins of the SPFH (band 7) domain superfamily may cluster to form membrane microdomains which may in turn recruit multiprotein complexes. These microdomains, in addition to being stable scaffolds, may also be dynamic units with their own regulatory functions. Prohibitin is a mitochondrial inner-membrane protein which may act as a chaperone for the stabilization of mitochondrial proteins. Human prohibitin forms a hetero-oligomeric complex with Bap-37 (prohibitin 2, an SPFH domain carrying homolog). This complex may protect non-assembled membrane proteins against proteolysis by the m-AAA protease. Prohibitin and Bap-37 yeast homologs have been implicated in yeast longevity and in the maintenance of mitochondrial morphology. |
| 173412 | PTZ00121 | 0.007 | 426 | 472 | 1114 | 1163 | MAEBL; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.69e-26 | 159 | 375 | 51 | 252 | |
| 5.04e-26 | 159 | 375 | 58 | 259 | |
| 1.90e-23 | 144 | 425 | 45 | 300 | |
| 1.90e-23 | 144 | 425 | 45 | 300 | |
| 1.90e-23 | 144 | 425 | 45 | 300 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.37e-24 | 144 | 425 | 45 | 300 | Beta-1,3-galactosyltransferase sqv-2 OS=Caenorhabditis elegans OX=6239 GN=sqv-2 PE=2 SV=1 |
|
| 4.26e-20 | 138 | 375 | 48 | 262 | Beta-1,3-galactosyltransferase 6 OS=Homo sapiens OX=9606 GN=B3GALT6 PE=1 SV=2 |
|
| 1.65e-17 | 155 | 410 | 47 | 282 | Beta-1,3-galactosyltransferase 6 OS=Mus musculus OX=10090 GN=B3galt6 PE=2 SV=1 |
|
| 1.05e-16 | 216 | 363 | 299 | 441 | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 OS=Danio rerio OX=7955 GN=b3galnt2 PE=2 SV=1 |
|
| 3.26e-16 | 216 | 425 | 294 | 484 | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 OS=Xenopus tropicalis OX=8364 GN=b3galnt2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999828 | 0.000180 |