You are browsing environment: FUNGIDB
CAZyme Information: AMAG_13248-t26_1-p1
You are here: Home > Sequence: AMAG_13248-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
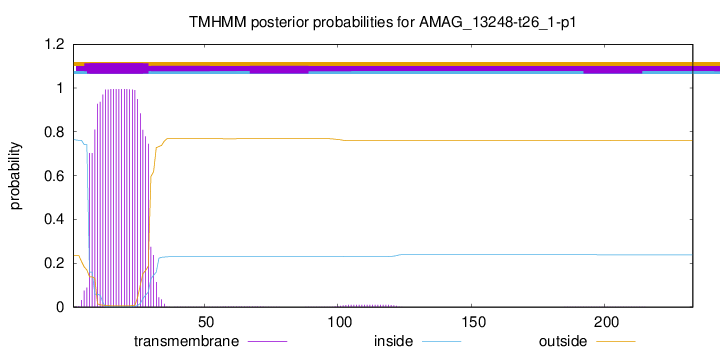
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_13248-t26_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:104 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 55 | 226 | 3.7e-54 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 1.64e-67 | 54 | 225 | 1 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.54e-68 | 44 | 228 | 42 | 227 | |
| 1.70e-67 | 51 | 233 | 57 | 240 | |
| 1.00e-66 | 44 | 228 | 42 | 227 | |
| 1.26e-66 | 45 | 228 | 43 | 227 | |
| 2.00e-66 | 45 | 228 | 45 | 229 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.06e-66 | 44 | 230 | 3 | 190 | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa [Thermopolyspora flexuosa] |
|
| 8.18e-66 | 44 | 228 | 5 | 190 | Chain A, Endo-1,4-beta-xylanase [Actinomycetia bacterium],7EO6_B Chain B, Endo-1,4-beta-xylanase [Actinomycetia bacterium] |
|
| 1.40e-65 | 45 | 228 | 45 | 230 | Crystal structure of XlnB2 [Streptomyces lividans],5EJ3_B Crystal structure of XlnB2 [Streptomyces lividans] |
|
| 1.60e-65 | 45 | 228 | 5 | 190 | Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],7DFM_B Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],7DFM_C Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],7DFM_D Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 [Streptomyces olivaceoviridis],7DFN_A Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with alpha-L-arabinofuranosyl xylotetraose [Streptomyces olivaceoviridis],7DFN_B Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with alpha-L-arabinofuranosyl xylotetraose [Streptomyces olivaceoviridis],7DFN_C Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with alpha-L-arabinofuranosyl xylotetraose [Streptomyces olivaceoviridis],7DFN_D Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with alpha-L-arabinofuranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_A Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_B Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_C Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_D Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_E Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_F Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_G Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_H Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis],7DFO_I Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with 4-O-methyl-alpha-D-glucuronopyranosyl xylotetraose [Streptomyces olivaceoviridis] |
|
| 2.82e-65 | 45 | 228 | 2 | 186 | 3D Structure of a thermophilic family GH11 xylanase from Thermobifida fusca [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.95e-64 | 39 | 228 | 49 | 239 | Endo-1,4-beta-xylanase B OS=Streptomyces sp. OX=1931 GN=xynBS9 PE=1 SV=1 |
|
| 1.45e-63 | 45 | 228 | 45 | 230 | Endo-1,4-beta-xylanase B OS=Streptomyces lividans OX=1916 GN=xlnB PE=1 SV=3 |
|
| 3.49e-59 | 44 | 228 | 52 | 239 | Endo-1,4-beta-xylanase C OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=xlnC PE=3 SV=1 |
|
| 1.55e-57 | 44 | 228 | 52 | 238 | Endo-1,4-beta-xylanase C OS=Streptomyces lividans OX=1916 GN=xlnC PE=1 SV=1 |
|
| 1.45e-56 | 45 | 230 | 44 | 230 | Bifunctional xylanase/deacetylase OS=Cellulomonas fimi OX=1708 GN=xynD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
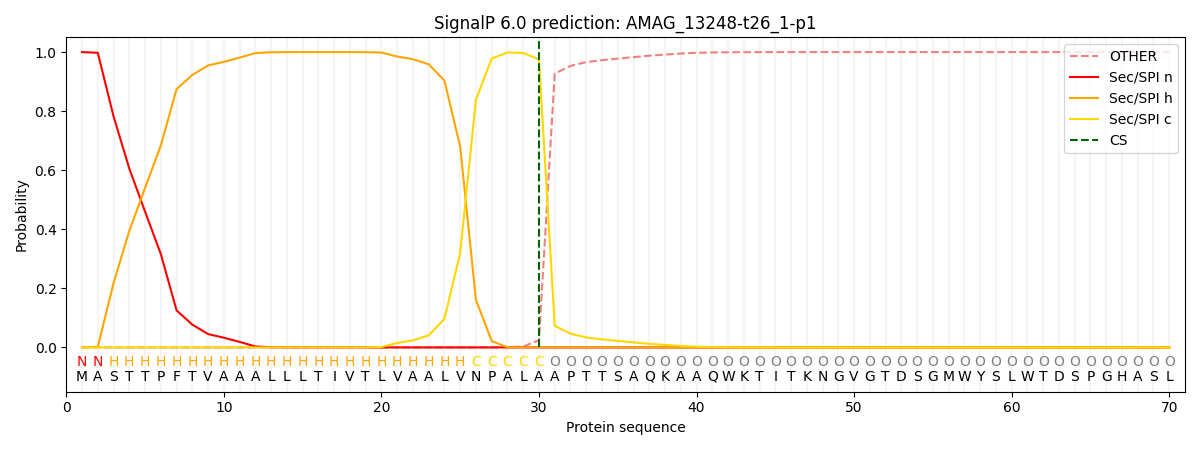
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000411 | 0.999580 | CS pos: 30-31. Pr: 0.9755 |