You are browsing environment: FUNGIDB
CAZyme Information: AMAG_12992-t26_1-p1
Basic Information
help
| Species |
Allomyces macrogynus
|
| Lineage |
Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus
|
| CAZyme ID |
AMAG_12992-t26_1-p1
|
| CAZy Family |
GT2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 582 |
GG745356|CGC3 |
61886.76 |
9.5400 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AmacrogynusATCC38327 |
19333 |
578462 |
535 |
18798
|
|
| Gene Location |
No EC number prediction in AMAG_12992-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH26 |
233 |
403 |
1.3e-20 |
0.40264026402640263 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 226609
|
ManB2 |
4.87e-06 |
229 |
399 |
161 |
297 |
Beta-mannanase [Carbohydrate transport and metabolism]. |
| 396639
|
Glyco_hydro_26 |
0.001 |
233 |
410 |
137 |
284 |
Glycosyl hydrolase family 26. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.22e-46 |
142 |
419 |
71 |
354 |
| 3.33e-46 |
140 |
408 |
48 |
319 |
| 1.70e-43 |
141 |
403 |
75 |
340 |
| 2.59e-43 |
140 |
365 |
48 |
266 |
| 3.14e-43 |
142 |
403 |
44 |
309 |
AMAG_12992-t26_1-p1 has no PDB hit.
AMAG_12992-t26_1-p1 has no Swissprot hit.
This protein is predicted as OTHER
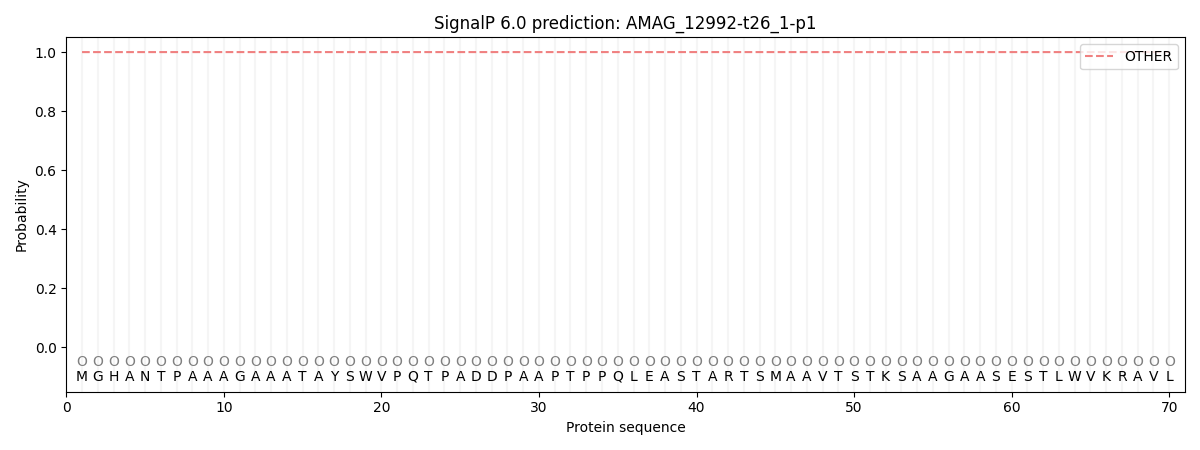
| Other |
SP_Sec_SPI |
CS Position |
| 1.000044 |
0.000003 |
|