You are browsing environment: FUNGIDB
CAZyme Information: AMAG_09045-t26_1-p1
You are here: Home > Sequence: AMAG_09045-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_09045-t26_1-p1 | |||||||||||
| CAZy Family | GH38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 21 | 177 | 5.3e-42 | 0.8481675392670157 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 237864 | PRK14950 | 0.002 | 220 | 276 | 377 | 431 | DNA polymerase III subunits gamma and tau; Provisional |
| 411607 | Chlamy_inclu_1 | 0.008 | 220 | 310 | 19 | 109 | inclusion-associated protein. Proteins of this family are inclusion-associated proteins in Chlamydia. It has been shown that protein CPj0783, which is identical to the HMM seed protein WP_010892266, was localized on Chlamydial inclusion. CPj0783 interacted with host Huntingtin-protein14, which may play an important role in disturbing the vesicle transport system to escape host lysosomal or autophagosomal degradation. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.94e-43 | 16 | 192 | 14 | 186 | |
| 1.89e-39 | 14 | 191 | 12 | 205 | |
| 2.59e-39 | 14 | 191 | 12 | 205 | |
| 2.59e-39 | 14 | 191 | 12 | 205 | |
| 2.78e-39 | 14 | 191 | 12 | 205 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.52e-17 | 21 | 162 | 1 | 153 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
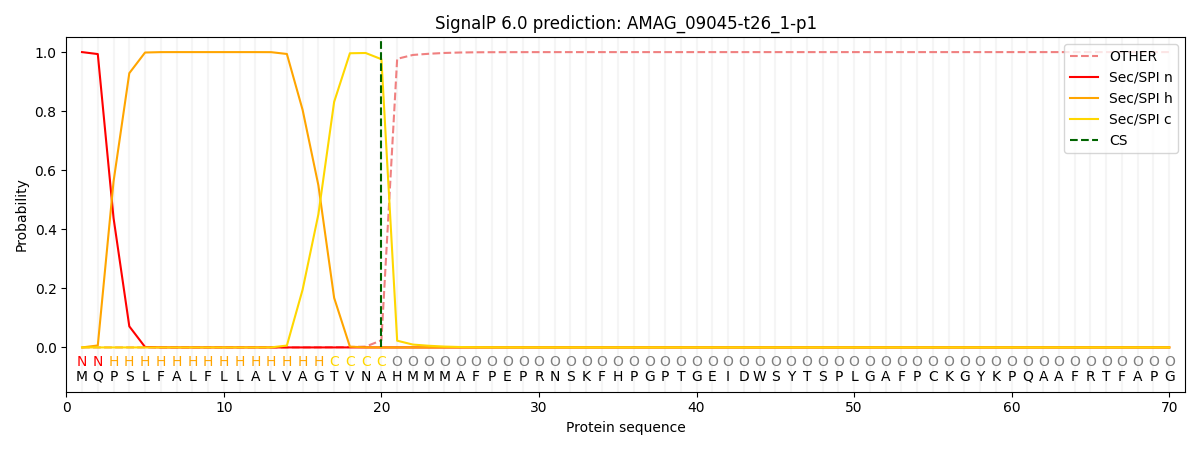
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000193 | 0.999789 | CS pos: 20-21. Pr: 0.9760 |

