You are browsing environment: FUNGIDB
CAZyme Information: AMAG_07617-t26_1-p1
You are here: Home > Sequence: AMAG_07617-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
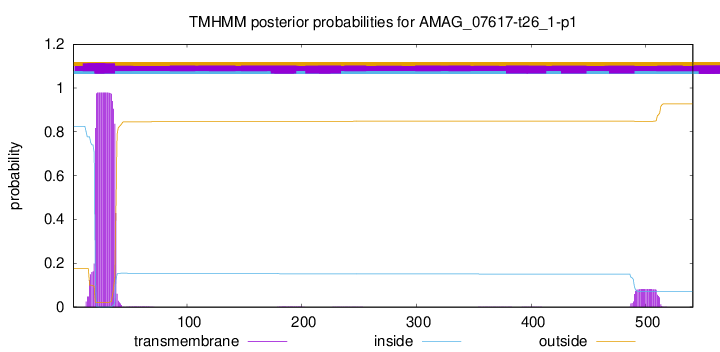
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_07617-t26_1-p1 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.149:9 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT31 | 209 | 404 | 1e-36 | 0.9635416666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 1.84e-18 | 209 | 407 | 3 | 194 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 215596 | PLN03133 | 2.98e-08 | 206 | 399 | 396 | 580 | beta-1,3-galactosyltransferase; Provisional |
| 178735 | PLN03193 | 3.26e-07 | 209 | 406 | 155 | 348 | beta-1,3-galactosyltransferase; Provisional |
| 396409 | TIL | 0.005 | 450 | 472 | 4 | 26 | Trypsin Inhibitor like cysteine rich domain. This family contains trypsin inhibitors as well as a domain found in many extracellular proteins. The domain typically contains ten cysteine residues that form five disulphide bonds. The cysteine residues that form the disulphide bonds are 1-7, 2-6, 3-5, 4-10 and 8-9. |
| 410995 | TIL | 0.009 | 450 | 469 | 4 | 23 | trypsin inhibitor-like cysteine rich domain. TIL (trypsin inhibitor-like) cysteine rich domains are found in smapins (small serine proteinase inhibitor), or Ascaris trypsin inhibitor (ATI)-like proteins, whose members include anticoagulant proteins, elastase inhibitors, trypsin inhibitors, thrombin inhibitors, and chymotrypsin inhibitors. The TIL domain is also found in some large modular glycoproteins, including the von Willebrand factor (VWF), mucin-6, mucin-19, and SCO-spondin, among others. The TIL domain is characterized by the presence of five disulfide bonds (two of which are located on either side of the reactive site) in a single small protein domain of 61-62 residues. The cysteine residues that form the disulfide bonds are linked in the pattern: cysteines 1-7, 2-6, 3-5, 4-10 and 8-9. TILs can occur as a single domain or in multiple tandem arrangements. The disulfide bonds account for the unusual resistance to proteolysis and heat denaturation of these proteins. Smapins possess an unusual fold and, with the exception of the reactive site, shows no similarity to other serine protease inhibitors. The serine protease inhibitors comprise a large family of molecules involved in inflammatory responses, blood clotting, and complement activation. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.40e-21 | 193 | 461 | 74 | 311 | |
| 1.16e-20 | 193 | 461 | 74 | 311 | |
| 1.56e-19 | 209 | 462 | 79 | 303 | |
| 3.13e-18 | 209 | 462 | 79 | 303 | |
| 3.84e-18 | 193 | 460 | 51 | 290 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.23e-17 | 257 | 460 | 303 | 488 | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 OS=Danio rerio OX=7955 GN=b3galnt2 PE=2 SV=1 |
|
| 4.83e-16 | 182 | 398 | 44 | 250 | Beta-1,3-galactosyltransferase 5 (Fragment) OS=Pan paniscus OX=9597 GN=B3GALT5 PE=3 SV=1 |
|
| 6.63e-16 | 255 | 460 | 296 | 483 | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 OS=Xenopus tropicalis OX=8364 GN=b3galnt2 PE=2 SV=1 |
|
| 8.32e-16 | 182 | 398 | 44 | 250 | Beta-1,3-galactosyltransferase 5 (Fragment) OS=Pan troglodytes OX=9598 GN=B3GALT5 PE=3 SV=1 |
|
| 9.86e-16 | 182 | 398 | 44 | 250 | Beta-1,3-galactosyltransferase 5 OS=Homo sapiens OX=9606 GN=B3GALT5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
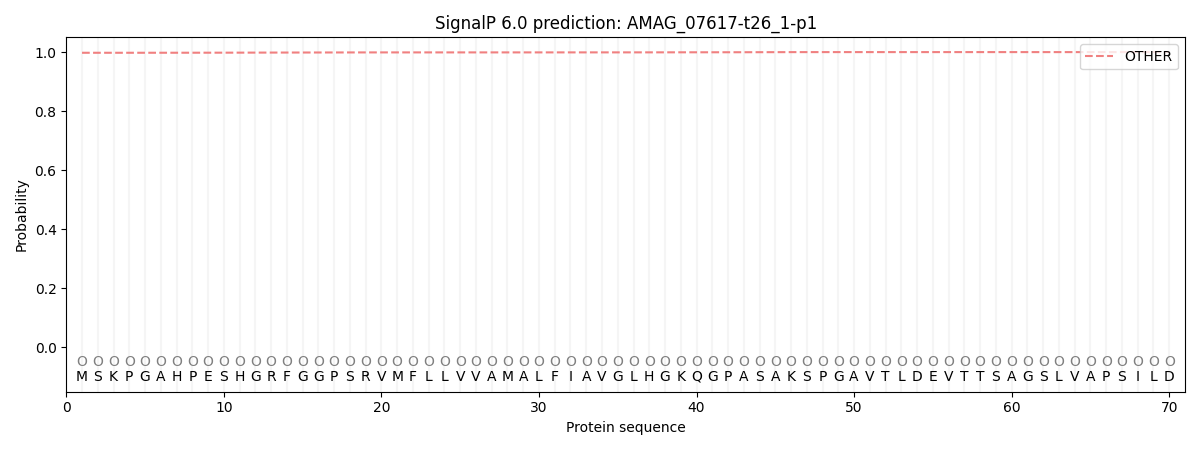
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.997938 | 0.002084 |