You are browsing environment: FUNGIDB
CAZyme Information: AMAG_06417-t26_1-p1
You are here: Home > Sequence: AMAG_06417-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
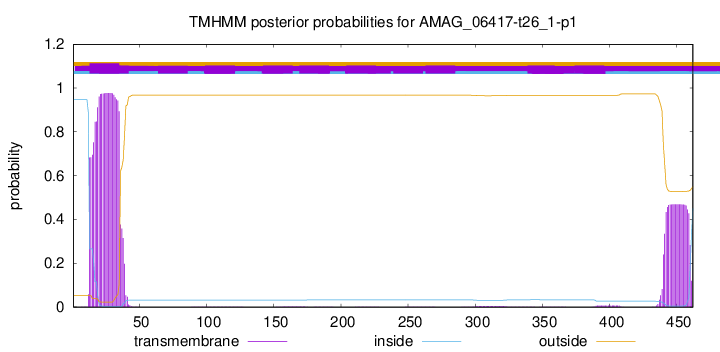
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_06417-t26_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 134 | 263 | 1.8e-18 | 0.8846153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 200575 | CE4_ClCDA_like | 9.62e-33 | 132 | 353 | 4 | 197 | Catalytic NodB homology domain of Colletotrichum lindemuthianum chitin deacetylase and similar proteins. This family is represented by the chitin deacetylase (endo-chitin de-N-acetylase, ClCDA, EC 3.5.1.41) from Colletotrichum lindemuthianum (also known as Glomerella lindemuthiana), which is a member of the carbohydrate esterase 4 (CE4) superfamily. ClCDA catalyzes the hydrolysis of N-acetamido groups of N-acetyl-D-glucosamine residues in chitin, converting it to chitosan in fungal cell walls. It consists of a single catalytic domain similar to the deformed (alpha/beta)8 barrel fold adopted by other CE4 esterases, which encompasses a mononuclear metalloenzyme employing a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine), to carry out acid/base catalysis. It possesses a highly conserved substrate-binding groove, with subtle alterations that influence substrate specificity and subsite affinity. Unlike its bacterial homologs, ClCDA contains two intramolecular disulfide bonds that may add stability to this secreted protein. The family also includes many uncharacterized deacetylases and hypothetical proteins mainly from eukaryotes, which show high sequence similarity to ClCDA. |
| 213022 | CE4_NodB_like_6s_7s | 3.15e-29 | 136 | 343 | 1 | 171 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| 200578 | CE4_CtAXE_like | 2.65e-21 | 137 | 345 | 2 | 170 | Catalytic NodB homology domain of Clostridium thermocellum acetylxylan esterase and its bacterial homologs. This family is represented by Clostridium thermocellum acetylxylan esterase (CtAXE, EC 3.1.1.72), a member of the carbohydrate esterase 4 (CE4) superfamily. CtAXE deacetylates O-acetylated xylan, a key component of plant cell walls. It shows no detectable activity on generic esterase substrates including para-nitrophenyl acetate. It is specific for sugar-based substrates and will precipitate acetylxylan, as a consequence of deacetylation. CtAXE is a monomeric protein containing a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold as other CE4 esterases. However, due to differences in the topography of the substrate-binding groove, the chemistry of the active center, and metal ion coordination, CtAXE has different metal ion preference and lacks activity on N-acetyl substrates. It is significantly activated by Co2+. Moreover, CtAXE displays distinctly different ligand coordination to the metal ion, utilizing an aspartate, a histidine, and four water molecules, as opposed to the conserved His-His-Asp zinc-binding triad of other CE4 esterases. |
| 200584 | CE4_GT2-like | 1.57e-19 | 136 | 358 | 1 | 196 | Catalytic NodB homology domain of uncharacterized bacterial glycosyl transferase, group 2-like family proteins. This family includes many uncharacterized bacterial proteins containing an N-terminal GH18 (glycosyl hydrolase, family 18) domain, a middle NodB-like homology domain, and a C-terminal GT2-like (glycosyl transferase group 2) domain. Although their biological function is unknown, members in this family contain a middle NodB homology domain that is similar to the catalytic domain of Streptococcus pneumoniae polysaccharide deacetylase PgdA (SpPgdA), an extracellular metal-dependent polysaccharide deacetylase with de-N-acetylase activity toward a hexamer of chitooligosaccharide N-acetylglucosamine, but not shorter chitooligosaccharides or a synthetic peptidoglycan tetrasaccharide. Like SpPgdA, this family is a member of the carbohydrate esterase 4 (CE4) superfamily. The presence of three domains suggests that members of this family may be multifunctional. |
| 200576 | CE4_MrCDA_like | 2.04e-17 | 136 | 327 | 1 | 162 | Catalytic NodB homology domain of Mucor rouxii chitin deacetylase and similar proteins. This family is represented by the chitin deacetylase (MrCDA, EC 3.5.1.41) encoded from the fungus Mucor rouxii (also known as Amylomyces rouxii). MrCDA is an acidic glycoprotein with a very stringent specificity for beta1-4-linked N-acetylglucosamine homopolymers. It requires at least four residues (chitotetraose) for catalysis, and can achieve extensive deacetylation on chitin polymers. MrCDA shows high sequence similarity to Colletotrichum lindemuthianum chitin deacetylase (endo-chitin de-N-acetylase, ClCDA), which consists of a single catalytic domain similar to the deformed (beta/alpha)8 barrel fold adopted by the carbohydrate esterase 4 (CE4) superfamily, which encompasses a mononuclear metalloenzyme employing a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. The family also includes some uncharacterized eukaryotic and bacterial homologs of MrCDA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.82e-22 | 121 | 358 | 18 | 222 | |
| 7.38e-20 | 126 | 363 | 29 | 241 | |
| 8.21e-20 | 129 | 358 | 32 | 229 | |
| 1.71e-19 | 129 | 364 | 38 | 243 | |
| 7.91e-19 | 129 | 360 | 32 | 229 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.64e-17 | 129 | 358 | 26 | 222 | Chain A, CHITIN DEACETYLASE [Aspergillus nidulans],2Y8U_B Chain B, CHITIN DEACETYLASE [Aspergillus nidulans] |
|
| 5.60e-16 | 129 | 275 | 33 | 166 | Chain A, Aspergillus niger contig An12c0130, genomic contig [Aspergillus niger CBS 513.88] |
|
| 8.08e-15 | 128 | 275 | 34 | 173 | Structure of the chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum [Colletotrichum lindemuthianum] |
|
| 3.26e-12 | 135 | 275 | 4 | 131 | Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate [Streptomyces lividans],2CC0_B Family 4 carbohydrate esterase from Streptomyces lividans in complex with acetate [Streptomyces lividans] |
|
| 3.46e-12 | 135 | 360 | 4 | 195 | Chain A, Glycoside Hydrolase, Family 11:clostridium Cellulosome Enzyme, Dockerin Type I:polysaccharide [Acetivibrio thermocellus],2C79_A Chain A, GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, DOCKERIN TYPE I:POLYSACCHARIDE [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.46e-20 | 129 | 358 | 32 | 229 | Chitin deacetylase OS=Pestalotiopsis sp. OX=36460 GN=CDA PE=1 SV=1 |
|
| 9.61e-17 | 129 | 358 | 33 | 229 | Chitin deacetylase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cda PE=1 SV=1 |
|
| 3.20e-15 | 120 | 366 | 153 | 371 | Chitin deacetylase OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=CDA PE=3 SV=1 |
|
| 2.07e-14 | 128 | 275 | 34 | 173 | Chitin deacetylase OS=Colletotrichum lindemuthianum OX=290576 GN=CDA PE=1 SV=1 |
|
| 4.07e-14 | 58 | 358 | 98 | 365 | Chitin deacetylase 1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=CDA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001395 | 0.998555 | CS pos: 41-42. Pr: 0.9383 |