You are browsing environment: FUNGIDB
CAZyme Information: AMAG_02762-t26_1-p1
You are here: Home > Sequence: AMAG_02762-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_02762-t26_1-p1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 25 | 387 | 5.2e-22 | 0.9527027027027027 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119365 | GH18_chitinase | 1.11e-21 | 26 | 381 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| 225862 | ChiA | 1.46e-19 | 16 | 381 | 30 | 423 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
| 214753 | Glyco_18 | 9.62e-19 | 25 | 381 | 1 | 334 | Glyco_18 domain. |
| 395573 | Glyco_hydro_18 | 2.69e-13 | 25 | 381 | 1 | 307 | Glycosyl hydrolases family 18. |
| 119358 | GH18_plant_chitinase_class_V | 1.81e-05 | 25 | 290 | 3 | 240 | The class V plant chitinases have a glycosyl hydrolase family 18 (GH18) domain, but lack the chitin-binding domain present in other GH18 enzymes. The GH18 domain of the class V chitinases has endochitinase activity in some cases and no catalytic activity in others. Included in this family is a lectin found in black locust (Robinia pseudoacacia) bark, which binds chitin but lacks chitinase activity. Also included is a chitinase-related receptor-like kinase (CHRK1) from tobacco (Nicotiana tabacum), with an N-terminal GH18 domain and a C-terminal kinase domain, which is thought to be part of a plant signaling pathway. The GH18 domain of CHRK1 is expressed extracellularly where it binds chitin but lacks chitinase activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.43e-17 | 22 | 317 | 236 | 524 | |
| 4.24e-15 | 16 | 381 | 65 | 431 | |
| 1.69e-14 | 25 | 346 | 189 | 501 | |
| 1.72e-14 | 20 | 309 | 110 | 399 | |
| 2.94e-14 | 25 | 384 | 34 | 394 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.49e-12 | 65 | 309 | 110 | 357 | Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis],6BT9_B Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis] |
|
| 3.74e-08 | 94 | 305 | 107 | 307 | Structure of Chitinase 42 from Trichoderma harzianum [Trichoderma harzianum] |
|
| 1.51e-07 | 94 | 305 | 107 | 307 | Chain A, Endochitinase 42 [Trichoderma harzianum],6YN4_A Chain A, Endochitinase 42 [Trichoderma harzianum],7AKQ_A Chain A, Endochitinase 42 [Trichoderma harzianum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.44e-08 | 98 | 315 | 112 | 330 | Endochitinase 46 OS=Trichoderma harzianum OX=5544 GN=chit46 PE=1 SV=1 |
|
| 4.44e-07 | 94 | 305 | 107 | 307 | Endochitinase 42 OS=Trichoderma harzianum OX=5544 GN=chit42 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
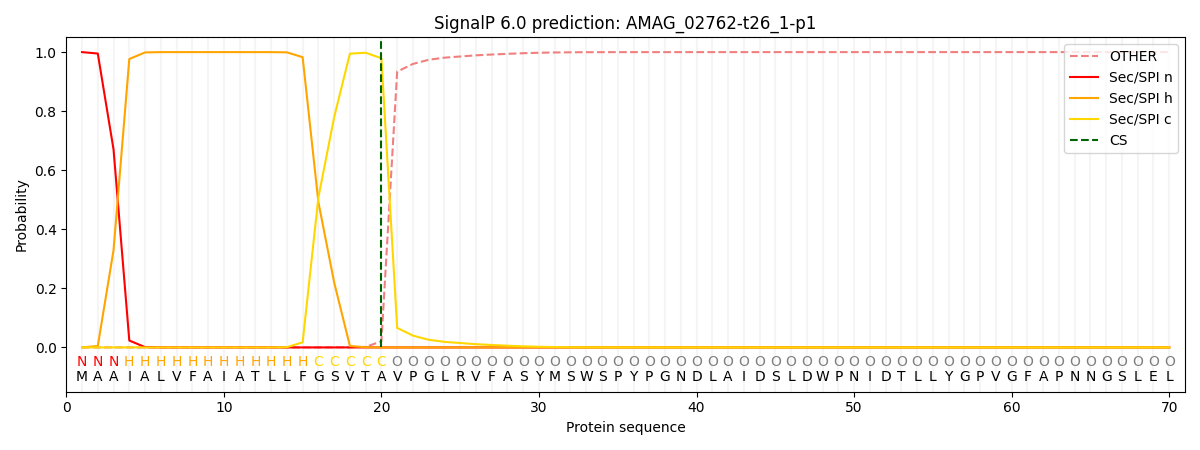
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000318 | 0.999652 | CS pos: 20-21. Pr: 0.9793 |

