You are browsing environment: FUNGIDB
CAZyme Information: AMAG_00758-t26_2-p1
You are here: Home > Sequence: AMAG_00758-t26_2-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_00758-t26_2-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | hypothetical protein, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.265:13 | 2.4.1.-:9 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT57 | 3 | 379 | 2.6e-107 | 0.7858627858627859 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397324 | Alg6_Alg8 | 3.76e-122 | 3 | 382 | 73 | 452 | ALG6, ALG8 glycosyltransferase family. N-linked (asparagine-linked) glycosylation of proteins is mediated by a highly conserved pathway in eukaryotes, in which a lipid (dolichol phosphate)-linked oligosaccharide is assembled at the endoplasmic reticulum membrane prior to the transfer of the oligosaccharide moiety to the target asparagine residues. This oligosaccharide is composed of Glc(3)Man(9)GlcNAc(2). The addition of the three glucose residues is the final series of steps in the synthesis of the oligosaccharide precursor. Alg6 transfers the first glucose residue, and Alg8 transfers the second one. In the human alg6 gene, a C->T transition, which causes Ala333 to be replaced with Val, has been identified as the cause of a congenital disorder of glycosylation, designated as type Ic OMIM:603147. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.32e-96 | 1 | 391 | 77 | 468 | |
| 2.88e-90 | 1 | 328 | 83 | 421 | |
| 2.88e-90 | 1 | 328 | 83 | 421 | |
| 1.11e-89 | 1 | 328 | 83 | 420 | |
| 1.14e-89 | 1 | 328 | 83 | 421 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.78e-16 | 17 | 288 | 159 | 410 | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc [Saccharomyces cerevisiae],6SNI_X Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.32e-84 | 1 | 319 | 83 | 407 | Dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ALG8 PE=3 SV=1 |
|
| 4.49e-83 | 1 | 319 | 83 | 408 | Dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=alg8 PE=3 SV=1 |
|
| 5.84e-83 | 1 | 338 | 83 | 421 | Dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Coccidioides immitis (strain RS) OX=246410 GN=ALG8 PE=3 SV=1 |
|
| 7.13e-82 | 1 | 339 | 83 | 431 | Dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-8 PE=3 SV=2 |
|
| 9.12e-81 | 1 | 335 | 80 | 414 | Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase OS=Drosophila melanogaster OX=7227 GN=xit PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000064 | 0.000000 |
TMHMM Annotations download full data without filtering help
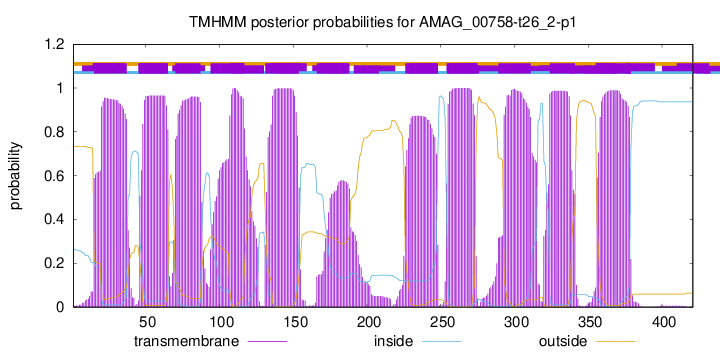
| Start | End |
|---|---|
| 15 | 37 |
| 46 | 65 |
| 70 | 87 |
| 94 | 116 |
| 131 | 153 |
| 166 | 188 |
| 226 | 248 |
| 255 | 274 |
| 289 | 311 |
| 324 | 341 |
| 356 | 378 |
