You are browsing environment: FUNGIDB
CAZyme Information: AMAG_00686-t26_1-p1
You are here: Home > Sequence: AMAG_00686-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
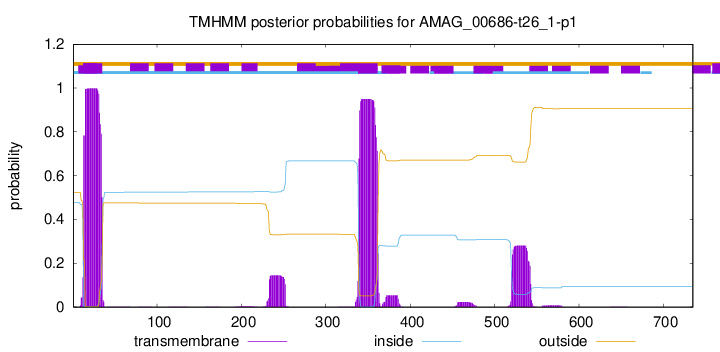
TMHMM annotations
Basic Information help
| Species | Allomyces macrogynus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Blastocladiomycota; Blastocladiomycetes; ; Blastocladiaceae; Allomyces; Allomyces macrogynus | |||||||||||
| CAZyme ID | AMAG_00686-t26_1-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.16:11 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 452 | 698 | 1.3e-19 | 0.4079696394686907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 367353 | Chitin_synth_2 | 1.72e-20 | 456 | 704 | 27 | 244 | Chitin synthase. Members of this family are fungal chitin synthase EC:2.4.1.16 enzymes. They catalyze chitin synthesis as follows: UDP-N-acetyl-D-glucosamine + {(1,4)-(N-acetyl-beta-D-glucosaminyl)}(N) <=> UDP + {(1,4)-(N-acetyl-beta-D-glucosaminyl)}(N+1). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.03e-18 | 450 | 706 | 1188 | 1399 | |
| 5.03e-18 | 450 | 706 | 1188 | 1399 | |
| 7.93e-17 | 456 | 703 | 1232 | 1435 | |
| 7.93e-17 | 456 | 703 | 1232 | 1435 | |
| 2.00e-16 | 456 | 704 | 410 | 614 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.17e-17 | 456 | 704 | 409 | 613 | Chitin synthase 6 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=CHS6 PE=3 SV=2 |
|
| 2.18e-16 | 456 | 704 | 1267 | 1471 | Chitin synthase 8 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=CHS8 PE=3 SV=1 |
|
| 1.93e-15 | 456 | 704 | 1255 | 1459 | Chitin synthase 5 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=CHS5 PE=2 SV=1 |
|
| 1.16e-14 | 456 | 697 | 508 | 705 | Chitin synthase 4 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=CHS4 PE=2 SV=2 |
|
| 2.43e-11 | 459 | 697 | 688 | 895 | Chitin synthase 4 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=CHS4 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
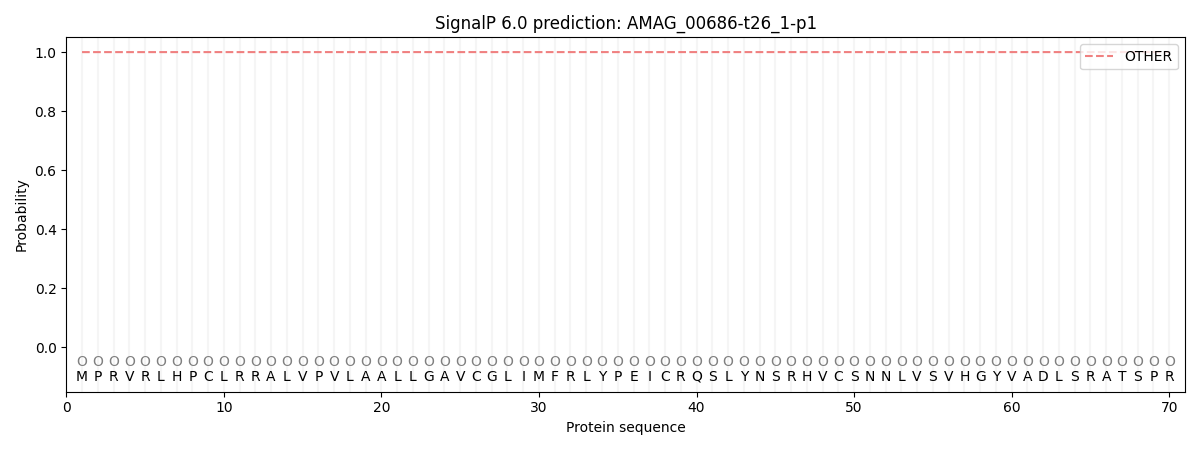
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999995 | 0.000025 |