You are browsing environment: FUNGIDB
CAZyme Information: ALNC14_043880:RNA-p1
You are here: Home > Sequence: ALNC14_043880:RNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Albugo laibachii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Albuginaceae; Albugo; Albugo laibachii | |||||||||||
| CAZyme ID | ALNC14_043880:RNA-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | predicted protein putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 128 | 393 | 4.2e-94 | 0.9961832061068703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178434 | PLN02841 | 4.73e-113 | 2 | 410 | 1 | 432 | GPI mannosyltransferase |
| 252941 | Mannosyl_trans | 3.93e-71 | 128 | 393 | 1 | 259 | Mannosyltransferase (PIG-M). PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars. It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. |
| 401508 | GT87 | 0.004 | 132 | 180 | 74 | 122 | Glycosyltransferase family 87. The enzymes in this family are glycosyltransferases. PimE is involved in phosphatidylinositol mannoside (PIM) synthesis, a major class of glycolipids in all mycobacteria. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of PIM. The family also includes alpha(1-->3) arabinofuranosyltransferase, invloved in the synthesis of of mycobacterial arabinogalactan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.33e-302 | 1 | 417 | 1 | 417 | |
| 4.97e-106 | 16 | 410 | 47 | 445 | |
| 1.26e-103 | 2 | 395 | 1 | 408 | |
| 2.76e-101 | 11 | 403 | 21 | 400 | |
| 5.48e-100 | 16 | 407 | 22 | 411 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.62e-103 | 7 | 397 | 27 | 431 | GPI mannosyltransferase 1 OS=Dictyostelium discoideum OX=44689 GN=pigm PE=3 SV=1 |
|
| 2.45e-97 | 16 | 399 | 25 | 412 | GPI mannosyltransferase 1 OS=Xenopus tropicalis OX=8364 GN=pigm PE=2 SV=1 |
|
| 2.77e-95 | 3 | 402 | 12 | 434 | GPI mannosyltransferase 1 OS=Arabidopsis thaliana OX=3702 GN=PIGM PE=2 SV=1 |
|
| 4.24e-95 | 13 | 394 | 16 | 403 | GPI mannosyltransferase 1 OS=Gallus gallus OX=9031 GN=PIGM PE=2 SV=2 |
|
| 1.39e-94 | 11 | 394 | 24 | 409 | GPI mannosyltransferase 1 OS=Rattus norvegicus OX=10116 GN=Pigm PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.106721 | 0.893246 | CS pos: 30-31. Pr: 0.7160 |
TMHMM Annotations download full data without filtering help
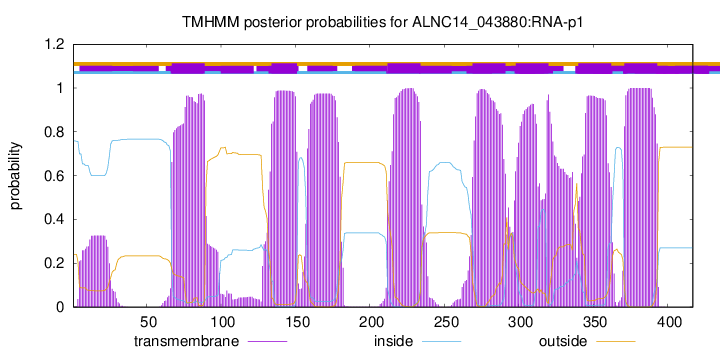
| Start | End |
|---|---|
| 67 | 89 |
| 134 | 151 |
| 212 | 234 |
| 269 | 291 |
| 298 | 320 |
| 340 | 362 |
| 371 | 393 |
